Where Evolutionary Tree Diagrams Come From: Evidence for Evolution (Part 9)
Where do “evolutionary tree” diagrams come from? Here are some underlying assumptions to remember next time you see an evolutionary tree presented as fact in a textbook.
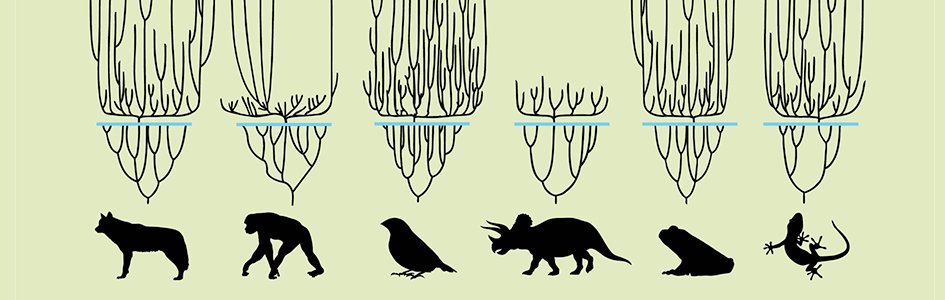
Whether walking through museums or flipping through textbooks, there’s a good chance you’ve come across an “evolutionary tree” image. Known as phylogenies, these branching diagrams supposedly show evolutionary relationships between fossils or living things. Those relationships may be presented as facts set in stone, like the fossils themselves. But are they really?
Describing organisms is observational science; however, speculating about their evolutionary past is historical science. To unearth some of the assumptions behind this historical science, let’s think about how researchers construct phylogenies. If you wanted to build an evolutionary tree, here are some steps you’d likely follow:
1. Choose Which Organisms to Include
When choosing organisms to map onto a tree, you’d normally already assume those organisms are evolutionarily related. It’s also common to include an outgroup—an organism you think is less closely related to the others. The outgroup provides a reference point for comparison, as you expect your analysis will show the other organisms are more similar to each other than to the outgroup.
2. Chose Which Characteristics to Compare
A basic evolutionary assumption states that, with some exceptions,1 the most similar organisms share the most recent common ancestor. So, constructing evolutionary trees requires scoring organisms’ similarities. The resulting diagram will show organisms with more overall similarities as being more closely related.
But which similarities will you analyze? Will you base your tree on physical similarities, genetic similarities, or both? Trees for fossil horses, for instance, are typically based on toe number and tooth type. Organisms alive today, however, can be compared using DNA or RNA sequence similarities.
There’s a catch, though. Shared ancestry cannot explain all similarities. Only homologies, or similarities assumed2 to be inherited from a common ancestor, “count” for constructing evolutionary trees. But, like Part 3 discussed, identifying homologies is not always straightforward and typically requires prior evolutionary assumptions.
3. Chose Which Parts of Those Characteristics to Compare
Many modern phylogenies compare homologous genes, known as orthologs. After identifying supposed orthologs,3 you assume that major differences between them were not present in the organisms’ supposed common ancestor. Instead, you assume those differences arose from DNA getting inserted or deleted after the organisms’ lineages separated. So, you ignore those differences and only compare parts of the sequences you assume came from the original ancestor. This is called aligning the genes.
Next, you can trim the aligned sequences by removing any parts deemed “low quality.”4 As a 2020 paper in Nature Reviews explained, “it is common to filter ambiguously aligned regions. Filtering can be based on ad hoc criteria regarding alignment quality such as gappyness and sequence similarity or by retaining only the alignment positions that are robust to changes in alignment parameters.” In other words, you get rid of remaining differences you don’t think should be included.
4. Choose How to Analyze Your Data
Once your data is ready, you can decide which statistical method you’ll use to analyze it. For instance, will you assume the most closely related organisms have the fewest differences overall, or the fewest evolutionary “steps” between them? Or will you base your analysis on the statistical likelihood of different evolutionary relationships, given your data?
None of these decisions is assumption-free. As one evolutionary researcher wrote, “molecular systematists of every provenance (those inclined to parsimony as well as those who prefer likelihood approaches) make biological and/or methodological assumptions about the nature of the evolutionary process.”5
Amassing Assumptions
All these assumptions affect which tree you end up with—assuming the type of evolution you’re trying to reconstruct is even possible. For example, it’s not uncommon for trees based on physical similarities to tell a different story from trees based on DNA similarities.6 That’s why entire groupings of supposedly related species had to be redefined after DNA sequencing became possible.
Even different gene comparisons often yield conflicting evolutionary trees, as a New Scientist article from 2009 observed.7 Entitled “Why Darwin Was Wrong About the Tree of Life,” the article also pointed out that different evolutionary trees based on RNA versus DNA don’t always match. People had hoped that comparing large samples of many genes would help clear up which evolutionary trees were most likely “correct.” But, like a paper from 2017 explained, “it has become clear that analyses of these large genomic data sets can also result in conflicting estimates of phylogeny.”7 This study found different evolutionary trees resulted from looking at many coding DNA sequences versus many non-coding DNA sequences.
Stories from different trees grow even more conflicted farther back along the supposed evolutionary timeline. As a 2014 paper observed,
For ancient clades, phylogenetic analysis of modern systematic datasets (many genes and many taxa) has taken two paths given currently available methods—supermatrix analysis or shortcut coalescence analysis. Each approach includes assumptions that are violated by empirical systematic data.9
It added that even when comparing longer DNA sequences, “wholesale conflicts among gene trees are apparent.”10
Freeing Fact from Fancy
Already, you can see why—as my own evolution professor emphasized—phylogenies represent hypotheses, not facts. They’re interpretations from historical science based on many layers of evolutionary assumptions. By keeping these assumptions in mind, you’ll be well prepared to think critically and biblically next time you see an evolutionary tree presented as fact in a museum or textbook.
Footnotes
- Not all similarities are considered the result of shared ancestry; see, for example, convergent evolution.
- When two species diverge within one kind of living thing (like dogs), the conclusion that their similarities are due to shared ancestry does not usually stem from historical assumption so much as direct observation; for instance, we can observe different dog species interbreeding.
- Orthologous genes are considered similar due to shared ancestry, while paralogous genes are considered similar due to duplication events.
- Paschalia Kapli, Ziheng Yang, and Maximilian J. Telford, “Phylogenetic tree building in the genomic age,” Nature Reviews Genetics, 21 (2020): 428–444.
- Edna Suárez-Díaza and Victor H.Anaya-Muñoz, “History, objectivity, and the construction of molecular phylogenies,” Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences 39, no 4 (2008): 451–468.
- Colin Patterson, David M. Williams, and Christopher J. Humphries, “Congruence between molecular and morphological phylogenies,” Annual review of ecology and systematics 24, no. 1: 153–188.
- Graham Lawton, “Why Darwin was wrong about the tree of life,” New Scientist 2692 (2009): 34–39.
- Sushma Reddy et al., “Why do phylogenomic data sets yield conflicting trees? Data type influences the avian tree of life more than taxon sampling” Systematic Biology, 66, no. 5 (2017): 857–879.
- John Gatesy and Mark S. Springer, “Phylogenetic analysis at deep timescales: unreliable gene trees, bypassed hidden support, and the coalescence/concatalescence conundrum,” Molecular phylogenetics and evolution 80 (2014): 231–266.
- Gatesy, “Phylogenetic analysis.”

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis
