
When Evolutionists Help Creationists Make Their Case, Part 3
Holding the Line on 40-year-old Secrets
On January 5, 1982, a federal judge handed creationists a legal loss—one with ripple effects for years to come. Judge Overton rejected an Arkansas act that stated, “Public schools within this State shall give balanced treatment to creation-science and to evolution-science.”1 This decision, and another subsequent one made by the US Supreme Court a few years later, have prevented the vast majority of recently trained scientists in the United States from being exposed to the technical young-earth creation (YEC) scientific literature as part of their mainstream scientific instruction.
Unsurprisingly, in light of this educational restriction, surveys show that over 98% of working PhD scientists accept evolution.2
Perhaps equally unsurprising is the way in which this statistic is used as an argument against the validity of creation science. For example, Richard Dawkins states:
Evolution is a fact, and [my] book will demonstrate it. No reputable scientist disputes it, and no unbiased reader will close the book doubting it.3 [emphasis added]
In one sense, Dawkins’ argument is circular. Evolutionists have actively sought to ban instruction in YEC views in the classroom. Then they turn around and use the results of this ban as evidence of the supposedly unscientific nature of YEC.
Testable Predictions
A second ripple effect of Judge Overton’s 1982 decision to reject creationism from the public school classroom arose from his reasoning. He first laid out his criteria for science:
The essential characteristics of science are:
- It is guided by natural law;
- It has to be explanatory by reference to natural law;
- It is testable against the empirical world;
- Its conclusions are tentative, i.e., are not necessarily the final word; and
- It is falsifiable.4 [emphases added]
He then declared:
creation science . . . fails to meet these essential characteristics.5
This criticism has been repeated by the evolutionary community for four decades. For example, famed paleontologist Niles Eldredge stated in 1982 that:
Creation-science isn’t science at all, nor have creation scientists managed to come up with even a single intellectually compelling, scientifically testable statement about the natural world. At least ninety-five percent of all of their reams of privately published books and pamphlets are devoted to an attack on conventional science. [Creationists] pose no testable hypotheses and make no predictions or observations worthy of the name.6 [emphases added]
More recently, a leading evolutionary textbook chided creationists using similar logic:
The most important feature of scientific hypotheses is that they are testable. . . . Scientists cannot test the hypothesis that an omnipotent God exists, or that He created anything, because we do not know what consistent patterns these hypotheses might predict.7 [emphasis theirs]
The Tables Turn
My book, Replacing Darwin, provides testable, falsifiable predictions that future experiments could reveal to be true or false.
Over the past decade, my colleagues and I have published several papers8 that tackle this challenge head-on in the realm of biology. In 2017, I published a book, Replacing Darwin,9 that provided exactly what the mainstream scientific and legal community has been demanding for 40 years—testable, falsifiable predictions that future experiments could reveal to be true or false.10
In response, the mainstream community has largely ignored it. In light of the past 40 years of history in the creation/evolution debate, I think I can understand why.
Countering YEC
However, a small group of young-earth creation (YEC) critics has attempted to rebut the latest arguments from YEC advocates, including the arguments in the papers that my colleagues and I have published. Their criticisms are significant for two reasons. First, they represent a group of YEC critics that are purportedly aware of the YEC technical scientific literature, yet still reject YEC science. If true, this would represent an argument against my thesis that scientists reject YEC because they are prevented from being exposed to it. Second, they have taken specific aim at the fact that we have published testable, falsifiable predictions.
I have previously documented11 how the leading opponents of YEC have recently resorted to misrepresentation to make their case. Furthermore, when one of the authors (David MacMillan, a former YEC) was confronted with the evidence of his professional scientific misconduct, he doubled down on his misrepresentations and added to them.12
MacMillan has now been made aware of both of these articles, and he has responded in an online forum (specific links to follow). His reactions give us a glimpse into just how important it is for evolutionists to hold the line of their claim that YEC scientists do not make testable predictions.
Initial Hints
In reaction to the latest of my published articles that document his misconduct, MacMillan repeated his misrepresentation of my work:
Jeanson seems extraordinarily angry. He can’t believe that we have read his work and still don’t agree with him.13
In fact, I explicitly stated the opposite in the article to which he’s responding:
The YEC answers to [MacMillan’s] questions have, of course, been published in the paper38 that MacMillan has repeatedly misrepresented. I’m sure he’d disagree with the answer. But this is a discussion for another day. The bigger issue is that MacMillan refuses to accurately represent what we’ve published. He’s free to disagree with our conclusions. But he cannot repeatedly lie about our position, and then call it science. It’s impossible to have a rational scientific discussion about a position that you refuse to accurately represent.14 [emphasis added]
Then MacMillan accused me of misrepresenting him:
We never claimed that Jeanson and Lisle agreed with our objections. We pointed out, using their own words, that they were at least tangentially aware of the problem with their model. “Jeanson and Lisle (2016) recognize yet another challenge to the postcreationist paradigm.” We couldn’t very well claim they agreed with the objections in a 2020 paper when we were citing a 2016 paper.15
Is MacMillan representing his own paper accurately? Here’s the full context for MacMillan et al.’s quote of—and reaction to—the Jeanson and Lisle (2016)16 paper:
In fact, Jeanson and Lisle (2016) recognize yet another challenge to the postcreationist [i.e., MacMillan et al.’s term for the modern young-earth creationist view] paradigm: they have identified no breaks or barriers to prevent the eventual emergence of species so different that, if compared, would certainly be viewed as representing different original kinds. They state: “hence, robust YEC explanations for the origin of a vast number of species must explain not only how genetic mechanisms produce many phenotypes, but also how these processes did not transform one kind into another.” Thus postcreationists, in their attempts to show scientific rigor, unwittingly uncover the very mechanisms that prove the opposite of their model. If they cannot identify a barrier to a modern species producing descendants which would by their method appear to descend from different original kinds, they cannot effectively argue that two species they currently consider to represent distinct kinds might not have originally diversified from a single ancestral clade.17
Clearly, MacMillan et al. use the quote of the Jeanson and Lisle (2016) paper as support of their position. And, as I showed in my previous articles,18 MacMillan et al. have falsely attributed to us a position to which we do not hold—a position which we explicitly reject in the very paper that they quote.
As hard as it is to believe, it also appears as if MacMillan thinks that I was honestly trying to argue that Jeanson and Lisle were, in 2016, projecting into the future, anticipating what critics would say in 2020. (MacMillan: “We couldn’t very well claim they agreed with the objections in a 2020 paper when we were citing a 2016 paper.”) I’m not sure anyone can infer this from what I wrote—except for someone with a history of misrepresentation, as I’ve already documented for MacMillan.
MacMillan then tried to avoid the main substance of my critique—by claiming that I avoided the main substance of their critique:
It’s amusing that, rather than engaging with the substance of our criticisms, Jeanson repeatedly focuses on this one tiny issue.19
Actually, the main substance of MacMillan’s criticisms has already been addressed. I responded directly in the first article in this series to this point:
Furthermore, many of the scientific points that they raise in the paper have already been addressed explicitly and implicitly. Conspicuously absent from the long list of creationist references at the end of their paper, are a reference to Replacing Darwin—the comprehensive written-for-critics explanation for the YEC origin of species—or a reference to a direct reply that I wrote to many of the points that Duff made previously on the BioLogos website, and repeated in the paper.20
In addition, if the issue that MacMillan calls “tiny” is so insignificant, why won’t he come clean about his misrepresentation? Why are we on the third round of exchanges, and yet he still refuses to accurately represent what we say? His actions betray the importance of this issue to him.
Testable Predictions Are Key
MacMillan then began to reveal one of the key reasons that he has so strongly resisted honest representations of what creationists have published.
MacMillan then began to reveal one of the key reasons that he has so strongly resisted honest representations of what creationists have published. His revelation explains much of his actions.
In my previous article, I exposed the logical incoherence of one of his defenses. He claimed that my views were scientifically untestable, despite having just published a paper purportedly showing the scientific implausibility of my views. In the online forum, MacMillan continued this contradictory pattern:
There are any number of nonsensical views which can be easily and readily disproven, yet ALSO lack falsifiable, verifiable, testable predictions … Jeanson’s failure to produce any verifiable, testable predictions to validate his model does not preclude someone like @evograd from pointing out all the ways Jeanson’s model fails …YEC is ultimately untestable because it is based on faith and commitment to ideology, not on evidence. Yet we can show with ease that any number of the models proposed by YEC fail miserably.21
Then MacMillan went to great lengths defending his view that my papers do not contain testable predictions (in light of what transpired later on the forum, I have highlighted some of MacMillan’s key claims):
The point Jeanson keeps trying (unsuccessfully) to belabor revolves around my 2016 statement that if he was really willing to advance his created heterozygosity model as valid, he should have used it to make verifiable predictions, but he does not. Jeanson protests that he DID make predictions! Of course, the predictions he made boiled down to “we predict new data will look like current data and therefore will be explicable by our model” which is not a meaningful prediction at all.
Could Jeanson make actual, meaningful, verifiable, testable, falsifiable predictions using his model? I don’t know. I doubt it. If he did, however, it would open his model up to falsification against the evidence, and that’s not what he’s about. It appears his “created heterozygosity” model can explain all conceivable data, which means it doesn’t actually explain anything at all.22
[N]one of the predictions he claims are proof of his model’s robustness are actually falsifiable.23 [emphases added]
A creationist on the same forum highlighted what would be evident to anyone who has read my papers or my book, Replacing Darwin:
Jeanson has made many scientifically testable predictions and he put them in his book.24
MacMillan doubled down:
No, he has not. Can you provide any example of a prediction he made, which predicts evidence NOT expected under the mainstream model, which could be confirmed or falsified by actual experiment?25
Later, MacMillan advanced his argument further:
If he wanted to show that his model was robust and useful, he could make predictions of mutagenic drift, base pair difference counts, and so forth in yet-unstudied species. These predictions would need to be different than the predictions of the mainstream model. They would need to be definite. They might take the form “My model predicst [sic] that once the genomes of (insert two species here) [sic] sequenced, we will find it has mtDNA BP differences on the order of 800, as opposed to the ~50 BP differences predicted by (insert actual mainstream research here).”26 [emphases added]
MacMillan made very clear what he meant by predictions:
We may be crossing purposes on the word “prediction”. I’m using it in the very limited sense of predicting a future discovery.27 [emphasis added]
MacMillan definitely was convinced that I made no testable predictions:
Despite Jeanson’s now lengthy protests to the contrary (four years running now), he does not make specific, defined testable predictions which do not align with the mainstream model.28 [emphasis added]
And to the YEC advocates, MacMillan offered a challenge:
If you disagree, please identify the predictions he makes, explain how they do not match what the mainstream model would predict, and explain what new evidences or discoveries would confirm or falsify the predictions.29 [emphases added]
MacMillan was adamant in his take on YEC views:
In order for a new model to demonstrate it has explanatory power, its originator needs to show that it deviates from the mainstream model in some measurable, real-world way…This is the problem with Jeanson’s so-called model. He’s just redefining a bunch of stuff and packing in fudge factors to create a hacked-together version of evolutionary biology that fits in his short timespan, but he fails to ever show that the predictions of this model deviate from mainstream expectations. He could try to show that, of course, but he has not done so. I have even provided examples of how he could do so, but I will not do his work for him. He has a Harvard PhD in biology; he should be capable of doing his own homework.30
Caught
A creationist began to point out to MacMillan that my published work does, in fact, make testable predictions.
A creationist began to point out to MacMillan that my published work does, in fact, make testable predictions. For example, from the Jeanson and Lisle (2016) paper:
For those species in which the vast majority of SNVs [i.e., single DNA letter variants] are inexplicable via constant rates of mutation over time, we predict that these SNVs will turn out to be functional, not non-functional or functionally neutral. In other words, we predict that these SNVs will participate in some way at the molecular level in the biology of each creature in a positive way. Rather than being “junk” DNA, molecular decoration, or harmful to the biology and function of an organism, we expect these variants to contribute to the development, expression, and/or operation of an organism’s traits.
In contrast, we expect most mutationally-derived variants (e.g., a small minority of the SNVs for most of the species and ‘kinds’ we examined) to be functionally neutral or slightly deleterious. Occasionally, some of these mutants might turn out to participate in the speciation process (e.g., see Lang et al. 2012) and therefore be viewed as “beneficial,” but we anticipate that the major effect of these variants will be to impede the normal function of the creature.
Contradictions between any of these predictions and future results would call into question aspects of our CHNP [i.e., Created Heterozygosity and Natural Processes] model and would cause us to reevaluate it.31
These predictions related to future experiments yet to be done; these predictions were specific; they were falsifiable; and they contrasted clearly with the expectations of evolution (e.g., see the referenced paper on evolutionary expectations for DNA function32 ).
Tellingly, these predictions also came from the paper33 that has been the subject of our three-part article series—it’s the one that MacMillan has repeatedly misrepresented.
Backtracking
How did MacMillan respond? By admitting his error? No. He said:
The problem is the adverbial clause “For those species in which the vast majority of SNVs are inexplicable via constant rates of mutation over time…”
It goes off the rails right at the beginning. This conditional allows him to pick and choose the data that fits. It’s circular … The prediction cannot possibly be “wrong” because it gives him the option to pick which data he wants to include.34 [emphases added]
If you’ve been following our exchange closely thus far, you probably won’t be surprised at the answer to his assertions. Yup, those “conditionals” were explicitly and clearly defined—in the paper itself. For example, for humans, we stated:
To clarify, within the world-wide human population, over 84 million total SNV [i.e., single DNA letter variants] sites have been identified (1000 Genomes Project Consortium et al. 2015). Rare variants, by definition, represent most of these 84 million sites. Since common variants would be present at identical sites in a variety of different individuals, common variants would constitute the minority of sites—they show up frequently but add little to the total number of different sites. By contrast, within each individual, only 3.5–4.3 million SNVs exist on average, and the vast majority of SNVs (>80%) within a single individual are common variants (1000 Genomes Project Consortium et al. 2012, 2015). Thus, within each individual, >80% of the nuclear SNVs (80% by the “common variant” criterion; >98% by Fig. 3B) are due to inheritance of alleles that arose via fiat creation during the Creation Week in Adam and Eve, and a small but significant minority of nuclear SNVs within an individual are due to mutations since Creation.35 [italics in original; bold emphasis added]
If this wasn’t clear enough, we showed exactly what we meant by “the vast majority of SNVs” that were “inexplicable via constant rates of mutation over time.” For example, look at the results on the right for humans—in particular, for Part (B) of that paper’s figure 3, shown below:
Fig. 3. Contradiction between mtDNA and nuclear DNA SNV clocks in humans.
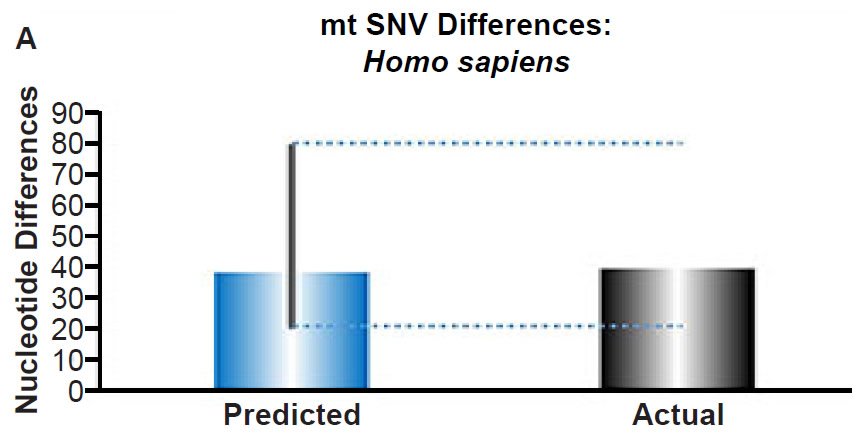
(A) Using the measured SNV mutation rate for the whole mitochondrial DNA (mtDNA) genome in non-African people groups, the number of mtDNA SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the current levels of mtDNA SNV differences in non-African people groups. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, the predicted number of differences overlapped the current differences among non-Africans. Adapted from Fig. 1 of Jeanson (2015b).
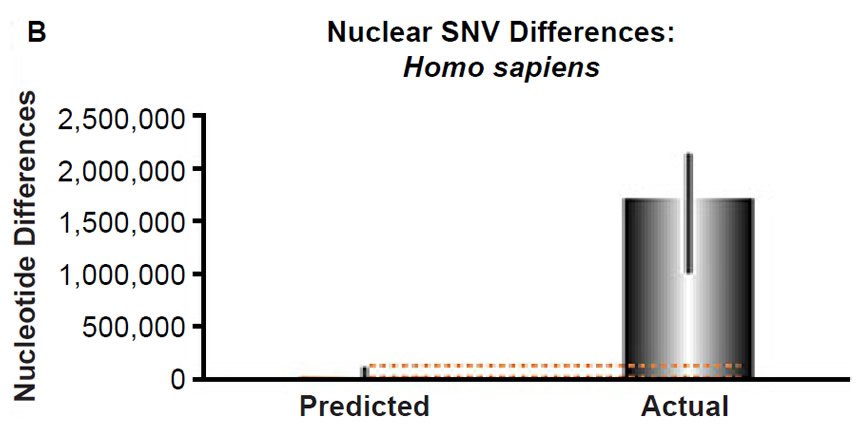
(B) Using the measured SNV mutation rate for the whole nuclear DNA genome in humans, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to heterozygosity estimates for individuals from several different non-African ethnic groups. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). For the “Actual” bar, the thick black lines represented the current range of reported nuclear DNA differences among non-African groups. As the dotted lines demonstrate, predictions clearly underestimated actual differences.
And for fruit flies, observe Part (B) of the Jeanson and Lisle, 2016, figure 4:
Fig. 4. Contradiction between mtDNA and nuclear DNA SNV clocks in Drosophila.
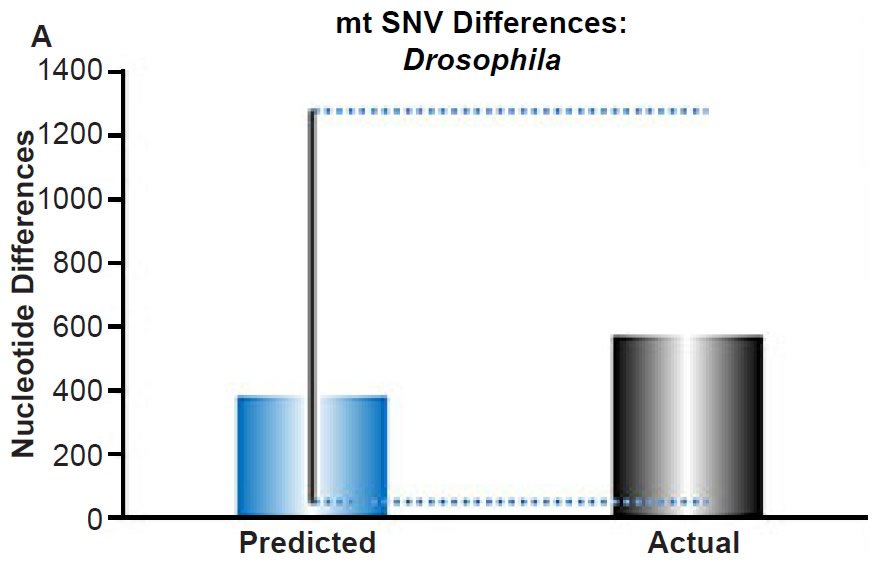
(A) Using the measured SNV mutation rate for the whole mitochondrial DNA (mtDNA) genome in Drosophila melanogaster, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the current levels of mtDNA SNV differences between D. melanogaster and D. simulans. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, the predicted number of differences captured the current levels of mtDNA differences between these two species. Adapted from Fig. 9 of Jeanson (2015a).
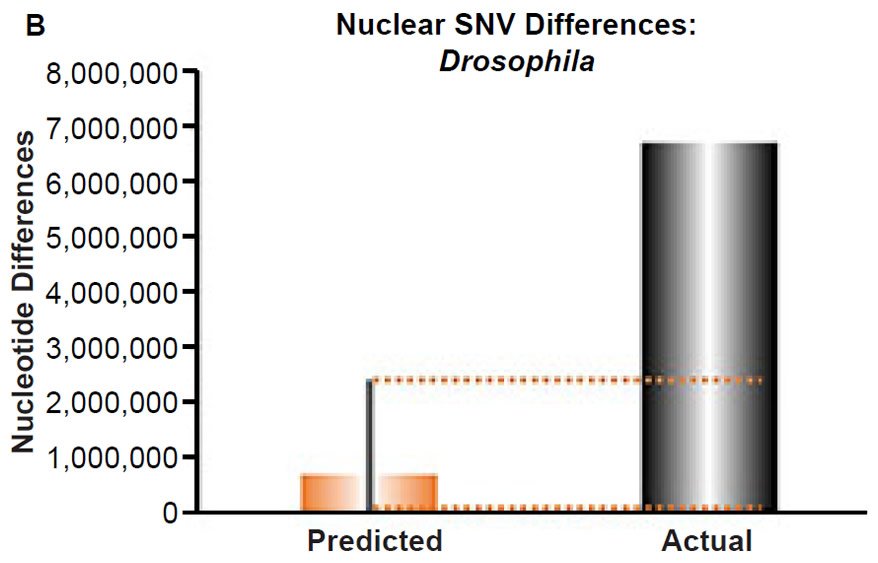
(B) Using the measured SNV mutation rate for the whole nuclear DNA genome in Drosophila melanogaster, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the current levels of nuclear SNV differences between D. melanogaster and D. simulans. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, predictions clearly underestimated actual differences.
And for water fleas, observe Part (B) of our figure 5:
Fig. 5. Contradiction between mtDNA and nuclear DNA SNV clocks in Daphnia pulex.
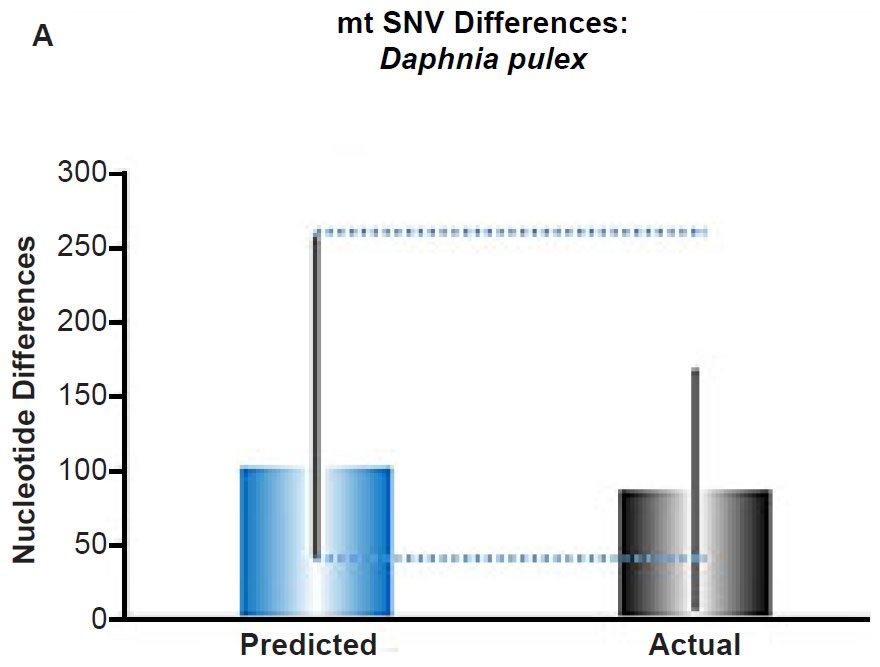
(A) Using the measured SNV mutation rate for the whole mitochondrial DNA (mtDNA) genome in Daphnia pulex, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the maximum SNV difference between Daphnia pulex individuals. The height of each colored bar represented the average (“Predicted” bar) or maximum (“Actual” bar) DNA difference, and the thick black lines represented the 95% confidence interval (“Predicted” bar). As the dotted lines demonstrate, the predicted number of differences captured the maximum current level of mtDNA differences among these individuals. Adapted from Fig. 10 of Jeanson (2015a).
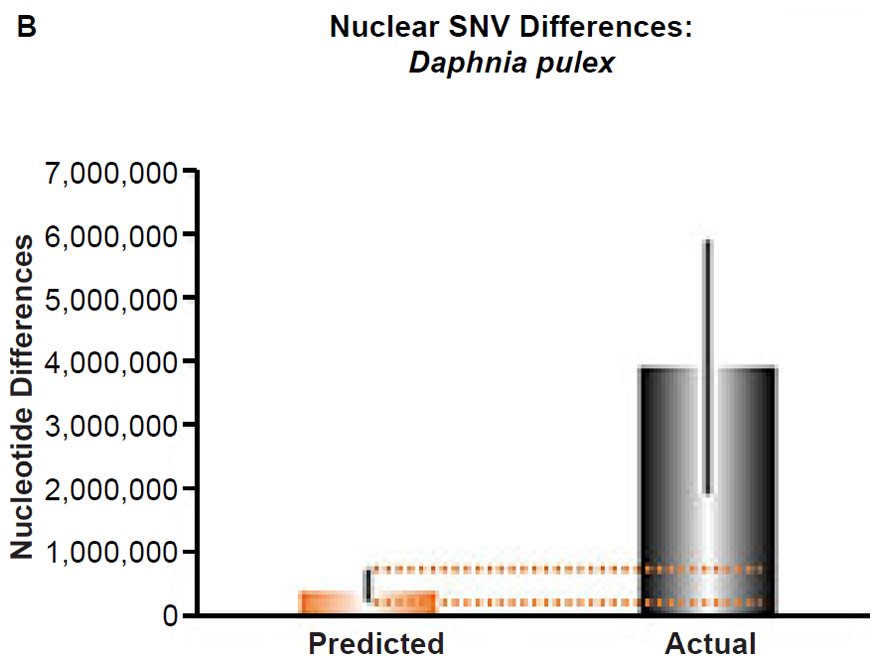
(B) Using the measured SNV mutation rate for the whole nuclear DNA genome in Daphnia pulex, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to an estimate of the current levels of nuclear SNV differences among D. pulex individuals. The height of each bar represented the average DNA difference. The thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar) or the reported range in heterozygosity levels among D. pulex individuals. As the dotted lines demonstrate, predictions clearly underestimated actual differences.
And for yeast, observe Part (B) of figure 6:
Fig. 6. Contradiction between mtDNA and nuclear DNA SNV clocks in yeast.
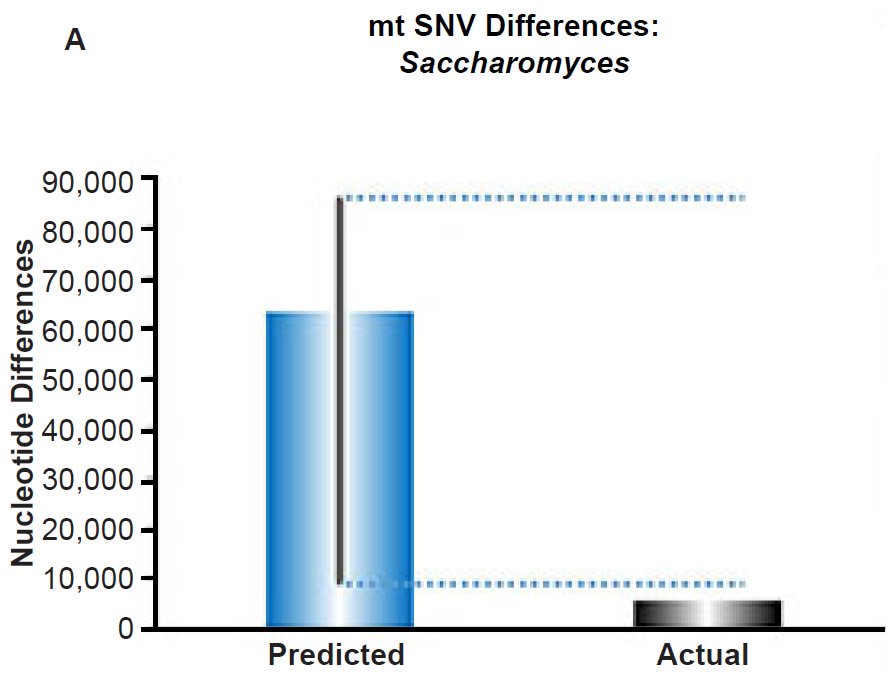
(A) Using the measured SNV mutation rate for the whole mitochondrial DNA (mtDNA) genome in Saccharomyces cerevisiae, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to an estimate of the current levels of mtDNA SNV differences between S. cerevisiae and S. paradoxus. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, the predicted number of differences over-predicted the current level of mtDNA differences between these species.
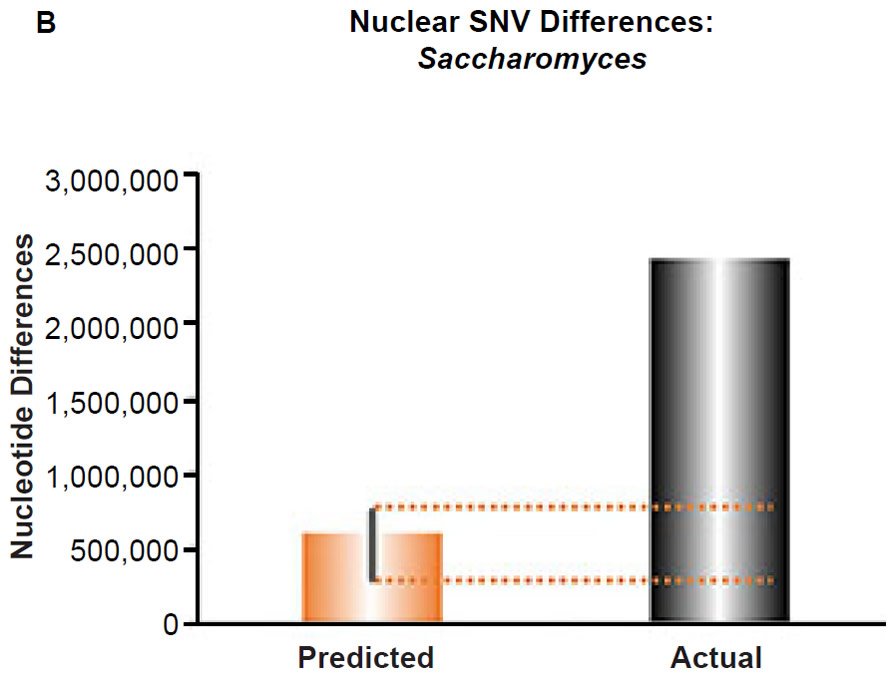
(B) Using the measured SNV mutation rate for the whole nuclear DNA genome in Saccharomyces cerevisiae, the number of SNV differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to an estimate of the current levels of nuclear SNV differences between S. cerevisiae and S. paradoxus. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, predictions clearly underestimated actual differences.
Check out how we defined “the vast majority of SNVs” that were “inexplicable via constant rates of mutation over time” for a specific plant species, for chimpanzees, and for mice:
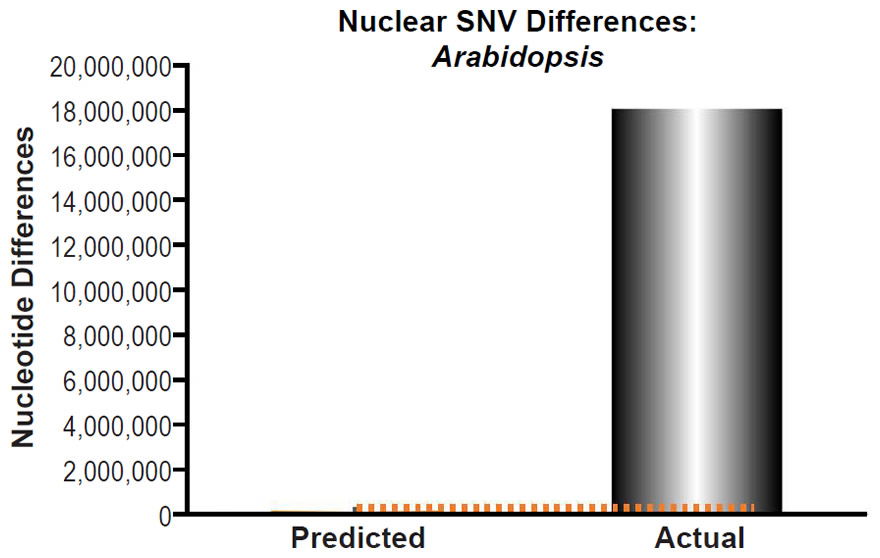
Fig. 12. Inability of mutations to explain Arabidopsis nuclear SNV differences. Using the measured SNV mutation rate for the whole nuclear DNA genome in ~homozygous individuals in Arabidopsis thaliana, the number of DNA differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the current levels of nuclear SNV differences between A. thaliana and A. lyrata. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, predictions clearly underestimated actual differences.
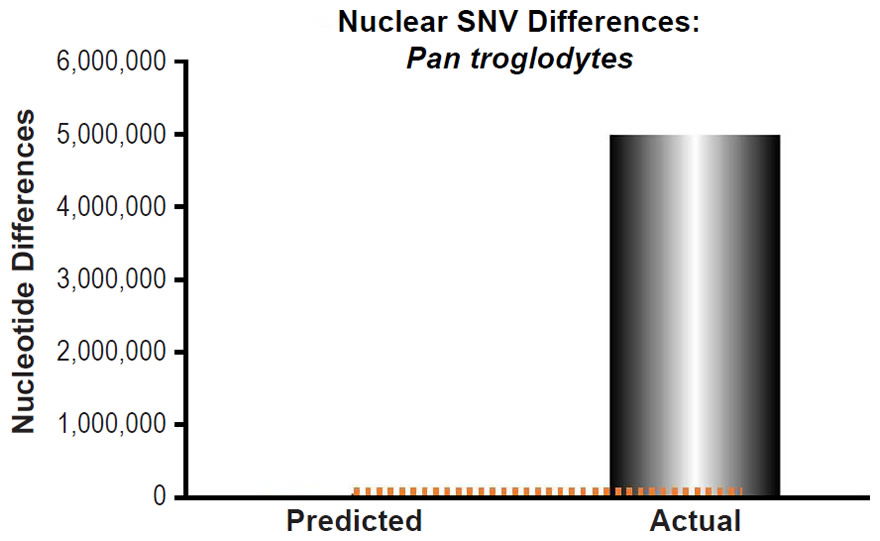
Fig. 14. Inability of mutations to explain chimpanzee nuclear SNV differences. Using the measured SNV mutation rate for the whole nuclear DNA genome in Pan troglodytes, the number of DNA differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the highest mean SNV per individual value among the published values for the various Pan troglodytes subspecies. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, predictions clearly underestimated actual differences.
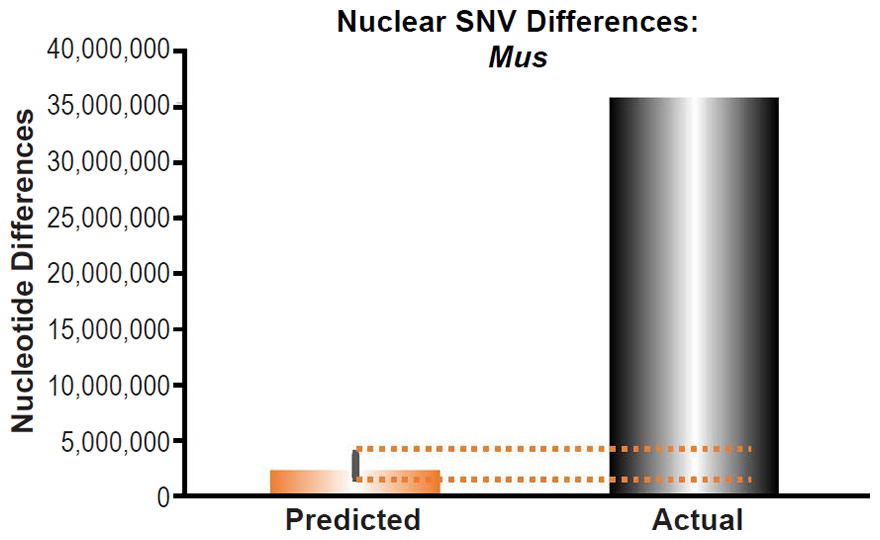
Fig. 15. Inability of mutations to explain mouse nuclear SNV differences. Using the measured SNV mutation rate for the whole nuclear DNA genome in Mus musculus, the number of DNA differences was predicted assuming a constant rate of DNA change over 6000 years. This prediction was compared to the number of SNV differences between laboratory mouse strains and a strain (SPRET/EiJ) derived from Mus spretus. The height of each bar represented the average DNA difference, and the thick black lines represented the range of predicted values given the reported error in the mutation rate and given the range of generation time estimates (“Predicted” bar). As the dotted lines demonstrate, predictions clearly underestimated actual differences.
For specific, detailed numbers, we published Supplemental Table 6 as part of the paper.36 Take a look. See if you can find anything not clearly, explicitly defined.
We could continue with more examples from the paper, but I think the point is clear in this example. Our paper contained explicit, specific definitions for the terms that MacMillan erroneously claimed were “conditional” and “circular,” and which give us “the option to pick which data he wants to include.”
MacMillan displayed the same behavior in a second example. He was confronted with another YEC statement that I published—a clear, specific, testable, falsifiable prediction that future experiments could reveal to be true or false—on human genetics:
I predict that the most diverse African groups will be found to mutate their mtDNA at a rate ~2-fold faster than reported here (e.g., at 0.2 to 0.3 mutations per genome per generation rather than 0.16 mutations per genome per generation).37 [emphases added]
MacMillan tried to rebut this claim in a similar way as the other:
Of course he will be able to choose that the “most diverse” African groups are the ones which fit his prediction, and he will say any groups that don’t fit his prediction “aren’t actually that diverse” if you look at more fudge factors he adds. A prediction that bakes in the ability to pick and choose data is no prediction at all.38 [emphases added]
Can you guess where the definition for “most diverse African groups” was found? Yup, one paragraph prior to the quote (and just prior to making the testable prediction) from my paper above:
The most diverse humans in the mtDNA databases are 123 base pairs apart (Kim and Schuster 2013)39 [emphases added]
This definition is explicit and referenced (i.e., the mainstream Kim and Schuster 2013 paper40); it’s given in terms (“base pairs”) that MacMillan demanded; and future observations can reveal whether this prediction is true or false. In other words, it’s a falsifiable claim.
Thus, even when contradictory data is held directly in front of MacMillan, he refuses to acknowledge it. He cannot concede that creationists actually make testable predictions. It’s almost like it’s impossible for him to relent on this point.
Why?
Endgame
For forty years, evolutionists have adamantly refused YEC science a place in the mainstream educational classroom. Because YEC science doesn’t make testable, falsifiable predictions, they say YEC science views cannot be taught in mainstream scientific classrooms. Consequently, having been shielded from YEC science claims in their educational instruction, the vast majority of mainstream scientists have endorsed evolution, not YEC. Then, evolutionists have used this fact to argue against the validity of YEC.
What happens when evolutionists admit that YEC scientists have published testable, falsifiable predictions? The legal case against YEC collapses.
Addendum
To date, MacMillan et al. are not the only critics of YEC science who have responded to my initial articles. Some have gone on the attack, attempting to expose YECs as equally guilty of scientific misconduct, and other supposed misdeeds. Yet, at present, none of them have acknowledged MacMillan et al.’s errors and misrepresentations. This silence—and reaction—is telling.
In the future, we’ll have some short blog posts to some of the more egregious claims (see Why Do So Many Reject Creation Science?).
Footnotes
- “McLean v. Arkansas Bd. of Ed., 529 F. Supp. 1255 (E.D. Ark. 1982),” January 5, 1982, https://law.justia.com/cases/federal/district-courts/FSupp/529/1255/2354824/.
- Pew Research Center, “An Elaboration of AAAS Scientists’ Views,” July 23, 2015, https://www.pewresearch.org/science/2015/07/23/an-elaboration-of-aaas-scientists-views/.
- Richard Dawkins, The Greatest Show on Earth (New York: Free Press, 2009), 9.
- “McLean v. Arkansas Bd. of Ed.,” 1982.
- Ibid.
- Niles Eldredge, The Monkey Business: A Scientist Looks at Creationism (New York: Washington Square Press, 1982), 80, 138.
- Douglas Futuyma and Mark Kirkpatrick, Evolution (Sunderland, MA: Sinauer Associates, Inc., 2017), p 578.
- Nathaniel T. Jeanson, Recent, functionally diverse origin for mitochondrial genes from ~2700 metazoan species, Answers Research Journal 6 (2013):467–501.
Nathaniel T. Jeanson, Mitochondrial DNA clocks imply linear speciation rates within “kinds.” Answers Research Journal 8 (2015):273–304.
Nathaniel T. Jeanson, A young-earth creation human mitochondrial DNA “clock”: Whole mitochondrial genome mutation rate confirms D-loop results. Answers Research Journal 8 (2015):375–378.
Nathaniel T. Jeanson, On the Origin of Human Mitochondrial DNA Differences, New Generation Time Data Both Suggest a Unified Young-Earth Creation Model and Challenge the Evolutionary Out-of-Africa Model. Answers Research Journal 9 (2016):123-130.
Nathaniel T. Jeanson and Jason Lisle, “On the Origin of Eukaryotic Species’ Genotypic and Phenotypic Diversity: Genetic Clocks, Population Growth Curves, and Comparative Nuclear Genome Analyses Suggest Created Heterozygosity in Combination with Natural Processes as a Major Mechanism,” Answers Research Journal 9 (2016):81–122.
Nathaniel T. Jeanson and Ashley D. Holland, Evidence for a Human Y Chromosome Molecular Clock: Pedigree-Based Mutation Rates Suggest a 4,500-Year History for Human Paternal Inheritance. Answers Research Journal 12 (2019):393–404.
Nathaniel T. Jeanson, Testing the Predictions of the Young-Earth Y Chromosome Molecular Clock: Population Growth Curves Confirm the Recent Origin of Human Y Chromosome Differences. Answers Research Journal 12 (2019):405–423.
Nathaniel T. Jeanson, 2020. Young-Earth Y Chromosome Clocks Confirm Known Post-Columbian Amerindian Population History and Suggest Pre-Columbian Population Replacement in the Americas. Answers Research Journal 13:23–33.
- Nathaniel T. Jeanson, Replacing Darwin: The New Origin of Species (Green Forest, AR: Master Books, 2017).
- In a sense, we were following in the footsteps of what the YEC geologists and physicists has done in their fields of science. For example, see the following:
Larry Vardiman, Andrew A. Snelling and E. F. Chaffin, eds. Radioisotopes and the Age of the Earth: Results of a Young-Earth Research Initiative. vol. 2 (El Cajon, CA: Institute for Creation Research and Chino Valley, AZ: Creation Research Society, 2005). - Nathaniel T. Jeanson, “When Evolutionists Help Creationists Make Their Case: A Shocking New Case of Professional Criticism Gone Bad,” June 30, 2020, https://answersingenesis.org/theory-of-evolution/when-evolutionists-help-creationists-make-their-case/.
- Nathaniel T. Jeanson, “When Evolutionists Help Creationists Make Their Case, Part 2,” July 7, 2020, https://answersingenesis.org/theory-of-evolution/when-evolutionists-help-creationists-make-their-case-part-2/.
- https://discourse.peacefulscience.org/t/jeanson-accuses-duff-of-misconduct-again/11024/40 .
- Jeanson, “When Evolutionists Help Creationists Make Their Case, Part 2”
- https://discourse.peacefulscience.org/t/jeanson-accuses-duff-of-misconduct-again/11024/40 .
- Jeanson and Lisle, “On the Origin.”
- R. Joel Duff, Thomas R. Beatman & David S. MacMillan III, “Dissent with modification: how postcreationism’s claim of hyperrapid speciation opposes yet embraces evolutionary theory,” Evolution: Education and Outreach 13, no. 9 (2020), https://doi.org/10.1186/s12052-020-00124-w.
- Jeanson, “When Evolutionists Help Creationists Make Their Case”; Jeanson, “When Evolutionists Help Creationists Make Their Case, Part 2.”
- https://discourse.peacefulscience.org/t/jeanson-accuses-duff-of-misconduct-again/11024/40.
- Jeanson, “When Evolutionists Help Creationists Make Their Case.”
- https://discourse.peacefulscience.org/t/jeanson-accuses-duff-of-misconduct-again/11024/40 .
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/5.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/11.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/13.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/14.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/19.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/22.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/150.
- Ibid.
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/157.
- Jeanson and Lisle, “On the Origin.”
Full Lang et al. 2012 reference as follows:
Michael Lang et al., “Mutations in the neverland Gene Turned Drosophila pachea into an Obligate Specialist Species” Science 337, no. 6102 (2012): 1658–1661. - Dan Graur et al., “On the Immortality of Television Sets: ‘Function’ in the Human Genome According to the Evolution-Free Gospel of ENCODE,” Genome Biology and Evolution 5, no. 3 (2013):578–590.
- Jeanson and Lisle, “On the Origin.”
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/176.
- Jeanson and Lisle, “On the Origin.”
Full 1000 Genomes Project Consortium et al. references as follows:
1000 Genomes Project Consortium, “An Integrated Map of Genetic Variation from 1,092 Human Genomes,” Nature 491, no. 7422 (2012): 56–65.
1000 Genomes Project Consortium, “A Global Reference for Human Genetic Variation,” Nature 526, no. 7571 (2015): 68–74. - Jeanson and Lisle, “On the Origin.”
- Nathaniel T. Jeanson, “A young-earth creation human mitochondrial DNA ‘clock’: Whole mitochondrial genome mutation rate confirms D-loop results.” Answers Research Journal 8 (2015):375–378. https://answersingenesis.org/genetics/mitochondrial-genome-mutation-rate/
- https://discourse.peacefulscience.org/t/comments-on-jeanson-accuses-duff-again/11069/176
- Jeanson, “A young-earth creation human mitochondrial DNA ‘clock’.”
- Hie Lim Kim and Stephan C. Schuster, “Poor Man’s 1000 Genome Project: Recent Human Population Expansion Confounds the Detection of Disease Alleles in 7,098 Complete Mitochondrial Genomes,” Frontiers in Genetics 4 (2013): 1–13, doi: 10.3389/fgene.2013.00013.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis






