Were Retroviruses Created Good?
Abstract
Retroviruses that are not normally present in healthy hosts are called exogenous viruses, while DNA sequences in cellular genomes that are homologous to retroviruses are called endogenous retroviruses (ERVs). While the belief that all ERVs are remnants of germline infection seems logical, there are also facts against the endogenization theory, such as xenotropic ERVs and essentiality of some ERVs in host physiology. Syncytins, products of the env gene of HERV-W and HERV-FRD, contribute to human placenta development. Similar genes are also found in mouse and sheep. Indeed, the sheep ERV genes have been shown essential for sheep reproduction. Furthermore, regulation of the human syncytin-1 gene involves a complex regulation network including both viral and host factors. Conclusion: While intact ERVs with positional polymorphism are likely germline copies of exogenous viruses, ERVs with fixed locations and conserved beneficial genes may have been incorporated into the host genome at the time of creation. Exogenous retroviruses may have been created to help the ERVs and to transfer useful genes between hosts.
And God saw every thing that he had made, and behold, it was very good. (Genesis 1:31)
Retroviruses: exogenous and endogenous
The main beneficial role of viruses known to date is the ecological role of horizontal gene transfer (DNA transduction) by bacteriophages.1,2 These viruses enable bacteria to share advantageous traits such as antibiotic resistance. The only family of viruses that are known to transduce genes among eukaryotic organisms (such as vertebrates) is that of the retroviruses. Retroviruses are viruses that have RNA as their genome but make DNA copies of it in the infected cell. They are similar to some bacteriophages (temperate phages) in that both insert their genetic material (DNA provirus) into host cell chromosome(s). This integration of viral and cellular DNA increases the chances of transduction of cellular genes adjacent to a provirus. Some retroviruses are known to transduce tumor genes (oncogenes) into the new host, which, while promoting the proliferation of the infected cell, often bring disaster to the organism.
While temperate phages only insert a single copy of the provirus into the host chromosome, retroviruses are allowed to insert multiple copies of proviruses into different sites of the same host genome. Integration of proviruses into the host’s germline cells (cells that give rise to eggs or sperm) will result in inherited retroviruses.3 On the other hand, the genomes of all vertebrates and humans harbor multiple copies of endogenous retroviruses (ERVs), DNA sequences that have genes and gene organizations homologous to those of retroviruses. Indeed, ERVs constitute about 8% of the human genome, a proportion much larger than the sum of all single-copy genes.4 While some ERVs are expressed and some even assembled into intracellular viral particles, most of them are deficient and are rarely transmitted horizontally. In view of this, retroviruses that are normally absent in healthy hosts are called exogenous retroviruses. Are ERVs degenerated germline copies of exogenous viruses which infected the host’s ancestors in history?
Evidences supporting the endogenization theory for the origin of ERVs include the following. (1) Modern exogenous viruses can infect the germline and be inherited like the host’s own genes.5 (2) Some endogenous viruses are replication-competent and infectious. When isolated murine leukemia viruses (MuLV, an endogenous mouse retrovirus) were inoculated into a new host, their proviruses were able to colonize the recipient genome.6 (3) Polymorphism (variation) of the chromosomal positions of an ERV among individuals of the same species suggests independent endogenization events. For instance, proviruses of the Koala retrovirus (KoRV) are found in different loci in different animals.7 (4) Allelic frequency polymorphism (variation in the frequency of finding a DNA sequence among populations) indicates recent endogenization. KoRV is present in koala in northern Australia, but absent in some animals in southern Australia.8 Similarly, HERV-K113, a provirus located on human chromosome 19p13.11, is rare among Caucasians and more prevalent among Africans, Asians, and Polynesians.9 (5) Negative correlation between degrees of positional polymorphism and sequence polymorphism conforms to the endogenization-degeneration hypothesis. ERVs with fixed chromosomal positions (less positional polymorphism) often demonstrate more sequence polymorphism with deleterious mutations and differences between the 5’ and 3’ long terminal repeats (LTRs), consistently pointing to the hypothesis that the virus infected an early ancestor and degenerated during its long history,10 while proviruses with varied locations such as KoRV are often intact and infectious, indicating recent or ongoing endogenization.
However, there are also interesting facts against the endogenization theory. (1) Endogenization of modern exogenous retroviruses is rarely observed in nature. (2) Most modern ERVs are not actively transposing (moving around or duplicating) in the host cell genome. At least all human ERVs appear fixed in numbers and positions; although some mouse ERVs are capable of expanding in the host genome. Are the human ERVs older, therefore more degenerated and less active? If the human race is younger than the murine race, as evolutionist biologists believe, there is no reason to suppose that the human ERVs are older than those of the mouse. (3) Xenotropic ERVs reside in cells that have no receptor for them. Instead, envelope (env) proteins of these ERVs bind receptors on cells of other animals.11 How did these ERVs get into the cell, if they were not built inside? It is no surprise to read speculations like this in Retroviruses, the “Bible” of retrovirology: “It is likely that xenotropic viruses originally inserted into the germ line in a host background that encoded their cognate receptor but that the functional xenotropic viral receptor allele was subsequently lost, probably under selective pressure from exogenous xenotropic viruses.”12 The term “exogenous xenotropic virus” is difficult to conceive, if not self-contradictory. (4) Essential beneficial functions of some ERVs and irreducibly complex coordination between ERVs and host DNA sequences argue against the possibility of historical acquisition of ERVs followed by positive selection (see below). (5) The existence of numerous solo LTRs in genomes suggests retroviral deletions, which may account for frequency polymorphism of some ERVs among populations. In other words, frequency polymorphism is a sign that the sequences are being lost, instead of being added on through endogenization.
Contributions of endogenous retroviruses to the host: syncytins
Retroelements including ERVs are normally suppressed from expression or transposition by extensive DNA methylation, RNA interference, heterochromatin formation, etc., to maintain genomic stability of the cell. Failure of the cell to control ERVs can lead to mutations or diseases.13,14,15 As a matter of fact, improper activation of endogenous retroviruses has been associated with many human cancers and autoimmune disorders.16 Complex interplay between retroelements and gene silencing mechanisms suggests ERVs are integral parts of the genome.
However, some of these genetic elements are expressed at certain stages of the host’s lifetime to the benefit of the host. Several instances of beneficial ERVs have been noticed. (1) Regulation, mainly activation, of neighboring (downstream) genes during embryonic development of the mouse.17 (2) Regulation of human genes expressed in the placenta (e.g. pleiotropin) and somatic tissues (e.g. apolipoprotein C1 in the liver and β-amylase in the salivary gland).18,19 (3) Immunomodulation, including induction of immunotolerance to self antigens and immunization against exogenous retroviruses.20,21 (4) The env protein of HERV-W and HERV-FRD serving as fusogenic factors (syncytins) during human trophoblast development.22,23 (5) Other roles such as mammalian tissue organization.24 Many of these genes are expressed during genome-wide DNA demethylation in gametogenesis (formation of eggs or sperm) and embryonic development, therefore are important for reproduction.
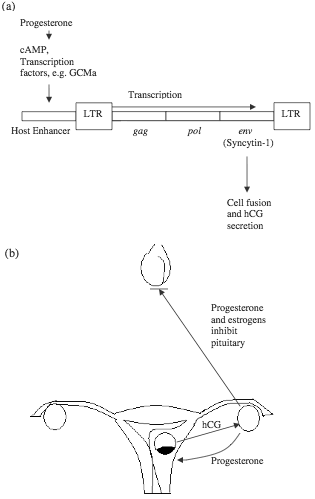
Figure 1. HERV-W is part of an irreducibly complex system in human reproduction. (a) HERV-W responds to ovarian progesterone and expresses syncytin-1, which mediates the formation of syncytiotrophoblasts and secretion of hCG. A number of transcription factors are involved in placenta-specific expression of syncytin-1. (b) Embryonic hCG and ovarian progesterone are interdependent during early pregnancy, while pituitary secretion of gonadotropins is inhibited by high levels of ovarian hormones.
There has been much investigation on the function and regulation of syncytins, especially syncytin-1, which is the env protein of HERV-W. Syncytin-1 is critical for the formation of syncytiotrophoblast (a layer of fused cells which invades the uterine wall and subsequently develops into parts of the placenta), and its secretion of human chorionic gonadotropin (hCG).25 Efficient tissue-specific expression of syncytin-1 requires cis-acting elements in both the 5’ LTR of HERV-W and host sequences (including an enhancer) upstream of the ERV.26 The host enhancer confers placental specificity to the expression of the syncytin gene, with binding sites responsive to cAMP/PKA and putative binding sites for AP-2, Sp-1, and GCMa, all of which are protein factors involved in embryonic development.27,28 GCMa is tissue-specific and has been shown to activate transcription of the syncytin-1 gene.29 Meanwhile, these transcription factors are under the control of host sex hormones such as progesterone. If the syncytins are indeed essential for human reproduction, they appear to be components of an irreducibly complex system that have to be created together to perform the intended function (figure 1).30 Recently, ERV env genes similar to syncytins have been shown to be essential for placenta development of sheep.31 Inhibition of these “viral” genes resulted in abortion in almost all animals. However, the authors of the report still believed in endogenization of these genes, and speculated that “it is likely that some of the host mechanisms governing the reproductive processes [before the endogenization (added for clarification)] may have been lost later during evolution.” The argument is similar to the explanation of xenotropic ERVs quoted above, both depending on “lost” elements which more likely never existed.
DNA sequences of Syncytin-1 are conserved in hominidae but appear degenerated in Old World Monkeys.32 While little is known about monkey genomes, complete sequences of the mouse genome are available. Muridae do not have genes orthologous to the HERV syncytins but have unrelated ERVs whose env genes code for similar proteins during placenta development.33 The case is similar in sheep.34 Evolutionary biologists call this phenomenon convergent evolution, speculating that hominidae, muridae, and bovidae were independently infected by different ERVs, which ended up to be harnessed in a similar way for a similar purpose in similar tissues, after the families separated from a common mammalian ancestor. To a creationist, it seems that the human, mouse, and sheep were created with a similar scheme regarding the formation of syncytiotrophoblast, but different viral materials were used.
Potential beneficial transduction by retroviruses: retrofection
In addition to the beneficial role of ERVs, exogenous retroviruses do transduce cellular nucleic acids other than oncogenes. While bacteriophages enter the host cell by nucleic acid translocation (only the viral DNA or RNA enters the cell), retroviruses penetrate by membrane fusion, allowing viral proteins and anything packaged in the virion to invade the cell. Cellular RNA molecules are frequently packaged in a retrovirus and transferred into other cells. Reverse transcription within the viral particle during the early stages of the viral life cycle is accompanied by high incidences of recombination, resulting in chimerical DNA, partly viral and partly cellular, which is subsequently integrated into chromosomes of the new host. This process is called retrofection. Retrofected cellular genes (retrogenes) have a common 16 nucleotide signature sequence.35 Viral LTR can serve as internal promoters for the expression of retrogenes. An example of a retrogene is the gene for the 7S L RNA, a component of the cytoplasmic signal recognition particle.36 Because laboratory models of retrofection involved human design, the natural role of retroviruses in horizontal gene transfer still awaits more solid evidence. Unfortunately, there has been no more report in this area since the early 1990s.
Conclusion
Rather than being added on during evolution by accidental endogenization of exogenous infectious agents, at least some ERVs must have been incorporated into the initial design of eukaryotic life. ERVs with fixed chromosomal positions are more likely integral parts of the host genome created in the host, while the more intact ERVs with positional polymorphism may be germline copies of exogenous viruses. The degenerative nature of mutation forbids the evolution of infectious viruses from ERVs. Exogenous viruses might have been created simultaneously with their endogenous counterparts during the Creation Week. Transmission and propagation of infectious retroviruses among the host population could have helped in maintenance of the endogenous viral sequences via recombination, in a way similar to recombinational DNA repair and modern gene therapy. (Indeed, retroviruses have been used as the classic vectors in gene therapy because of their ability to integrate into host chromosomes.) Interactions between endogenous and exogenous retroviruses may have been perfectly regulated at the time of creation. Additionally, infectious retroviruses may also have played a role in horizontal gene transfer which was ecologically beneficial. Insertional mutagenesis, formation of v-onc genes, as well as oncogene transduction, are rather dysregulated biological events performed by retroviruses after the Fall, or even after the Flood, when God decided to shorten the human life span. For similar reasons, we do not expect germline infection of modern exogenous viruses to bring benefits to the host, whether on the level of an individual or on the level of a species.
Glossary
Bacteriophage: a virus that infects bacteria; also called phage.
Cis-acting element: DNA sequences adjacent to a gene, which contain target sites for transcription factors made of protein.
DNA methylation and demethylation: addition and removal of a methyl (-CH3) group on carbon number 5 of cytosine. Methylated genes are typically suppressed while demethylation is associated with activation.
Endogenous retrovirus: cellular DNA sequences that have genes and gene organizations similar to that of infectious retroviruses.
Enhancer: cis-acting element with concentrated binding sites for multiple transcription factors. An enhancer upregulates the expression of the neighboring gene and is indeed essential for many genes.
Exogenous retroviruses: infectious retroviruses that are normally absent in healthy hosts.
Gametogenesis: formation of eggs or sperm in the ovaries or testes.
Genome: all the genes of an organism.
Irreducible complexity: “a single system composed of several well-matched, interacting parts that contribute to the basic function, wherein the removal of any one of the parts causes the system to effectively cease functioning.”37
Long terminal repeat (LTR): identical DNA sequences at both ends of a provirus.
Mutagenesis: creation of mutations.
Oncogenes: genes that cause tumor, oftentimes cancer.
Polymorphism (genetic polymorphism): variation of DNA sequences at a certain position of the genome.
Provirus: a DNA copy of the viral genome (which may be DNA or RNA in viral particles) that is integrated into the host chromosome.
Replication: reproduction of a virus in the host cell.
Retroelement: also retroposon, genetic elements that can change positions on chromosomes via an RNA copy. Include endogenous viruses and other simpler repetitive DNA sequences.
Retrofection: a retrovirus packages cellular nucleic acid molecules and carries them into a new host, resulting in horizontal transfer of genetic information.
Syncytiotrophoblast: outer layer of embryo, developed by fusion of single cells into one giant syncytium containing multiple nuclei. It is called chorion later in development. Branches grow out of it that spread within the uterine wall, eventually forming the placenta.
Temperate phage: a bacteriophage that integrates its genome into the host chromosome. It may remain in the cell without killing it for generations.
Transcription factor: protein factor that binds to cis-acting DNA elements to regulate the expression of a gene.
Transduction: a virus carries part of the host genome into another cell. It is analogous to a mosquito transferring the malaria parasite among people.
Transposition: change in position of a DNA sequence within the genome, by cut-pasting or copy-pasting.
Xenotropism: the inability to infect cells of the species where an endogenous virus is found, and ability to infect cells of other species.
About the author
Dr. Yingguang Liu earned his Ph.D. in Biological Sciences from Ohio University. He currently serves as an associate professor at Maranatha Baptist Bible College.
Footnotes
- Bergman, J., Did God make pathogenic viruses? Creation Ex Nihilo Technical Journal 13: 115-125, 1999.
- Chibani-Chennoufi, S., Bruttin, A., Dillmann, M.L., and Brussow, H., Phage-host interaction: an ecological perspective, Journal of bacteriology 186: 3677-3686, 2004.
- Jaenisch, R.F., Germline integration and Mendelian transmission of the exogenous Moloney leukemia virus, Proceedings of the National Academy of Sciences, USA 73: 1260-1264, 1976.
- Lewin, B., Genes VIII, Pearson Education, Inc., Upper Saddle River, NJ., pp. 505, 2004.
- Jaenisch, R.F., Germline integration and Mendelian transmission of the exogenous Moloney leukemia virus, Proceedings of the National Academy of Sciences, USA 73: 1260-1264, 1976.
- Lock, L.F., Keshet, E., Gilbert, D.J., Jenkins, N.A., and Copeland, N.G., Studies of the mechanism of spontaneous germline ecotropic provirus acquisition in mice, The EMBO Journal 7: 4169-4177, 1988.
- Tarlinton, R.E., Meers, J., and Young, P.R., Retroviral invasion of the koala genome, Nature 442:79-81, 2006.
- Tarlinton, R.E., Meers, J., and Young, P.R., Retroviral invasion of the koala genome, Nature 442:79-81, 2006.
- Turner, G., Barbuscu, M., Su, M. Jensen-Seaman, M.I., Kidd, K.K., and Lenz, J., Insertional polymorphisms of full-length endogenous retroviruses in humans, Current biology 11:1531-1535, 2001.
- Coffin, J.M., Retroviridae: the viruses and their replication; in: Field, B.N., Knipe, D.M., Howley, P.M., Chanock, R.M., Melnick, J.L., Monath, T.P., Roizman, B., and Straus, S. E. (eds.), Fields Virology, 3rd ed, Lippincott-Raven Publishers, Philadelphia, PA, pp. 1767-1847, 1996.
- Coffin, J.M., Retroviridae: the viruses and their replication; in: Field, B.N., Knipe, D.M., Howley, P.M., Chanock, R.M., Melnick, J.L., Monath, T.P., Roizman, B., and Straus, S. E. (eds.), Fields Virology, 3rd ed, Lippincott-Raven Publishers, Philadelphia, PA, pp. 1767-1847, 1996.
- Coffin, J.M., Hughes, S.H., and Varmus, H.E., Retroviruses, Cold Spring Harbor Laboratories Press, Plainview, Plainview, NY., p. 77, 1997.
- Coffin, J.M., Retroviridae: the viruses and their replication; in: Field, B.N., Knipe, D.M., Howley, P.M., Chanock, R.M., Melnick, J.L., Monath, T.P., Roizman, B., and Straus, S. E. (eds.), Fields Virology, 3rd ed, Lippincott-Raven Publishers, Philadelphia, PA, pp. 1767-1847, 1996.
- Druker, R. and Whitelaw E., Retrotransposon-derived elements in the mammalian genome: a potential source of disease, Journal of Inherited Metabolic Disease 27: 319-330, 2004.
- Schulz, W.A., Seinhoff, C., and Flori, A.R., Methylation of endogenous retroelements in health and disease, Current Topics in Microbiology and Immunology 310: 211-250, 2006.
- Schulz, W.A., Seinhoff, C., and Flori, A.R., Methylation of endogenous retroelements in health and disease, Current Topics in Microbiology and Immunology 310: 211-250, 2006.
- Peaston A.E., Evsikon, A.V., Graber, J.H., de Vries, W.N., Holbrook, A.E., Solter, D., and Knowles, B.B., Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos, Developmental cell 7:597-606, 2004.
- Coffin, J.M., Retroviridae: the viruses and their replication; in: Field, B.N., Knipe, D.M., Howley, P.M., Chanock, R.M., Melnick, J.L., Monath, T.P., Roizman, B., and Straus, S. E. (eds.), Fields Virology, 3rd ed, Lippincott-Raven Publishers, Philadelphia, PA, pp. 1767-1847, 1996.
- Bannert, N. and Kurth, R., Retroelements and the human genome: New perspectives on an old relation, Proceedings of the National Academy of Science, USA 101: 14572-14579, 2004.
- Coffin, J.M., Retroviridae: the viruses and their replication; in: Field, B.N., Knipe, D.M., Howley, P.M., Chanock, R.M., Melnick, J.L., Monath, T.P., Roizman, B., and Straus, S. E. (eds.), Fields Virology, 3rd ed, Lippincott-Raven Publishers, Philadelphia, PA, pp. 1767-1847, 1996.
- Bannert, N. and Kurth, R., Retroelements and the human genome: New perspectives on an old relation, Proceedings of the National Academy of Science, USA 101: 14572-14579, 2004.
- Mallet, F., Oliver, B., Prudhomme, S., Cheynet, V., Oriol, G., Bonnaud, B., Lucotte, G., Duret, L., and Mandrand, B., The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology, Proceedings of the National Academy of Sciences, USA 101: 1731-1736, 2004.
- Frendo, J.L., Olivier, D., Cheynet, V., Blond, J.L., Bouton, O., Vidaud, M., Rabreau, M., Evain-Brion, D., and Mallet, F., Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation, Molecular and Cellular Biology 23:3566-3574, 2003.
- Matsui, T., Kinoshita-Ida, Y., Hayashi-Kisumi, F., Hata, M., Matsubara, K., Chiba, M., Katahira-Tayama, S., Morita, K., Miyachi, Y., and Tsukita, S., Mouse homologue of skin-specific retroviral-like aspartic protease (SASPase) involved in wrinkle formation, The Journal of Biological Chemistry Jul 12; [Epub ahead of print], 2006.
- Frendo, J.L., Olivier, D., Cheynet, V., Blond, J.L., Bouton, O., Vidaud, M., Rabreau, M., Evain-Brion, D., and Mallet, F., Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation, Molecular and Cellular Biology 23:3566-3574, 2003.
- Prudhomme, S., Oriol, G, and Mallet, F., A retroviral promoter and a cellular enhancer define a bipartite element which controls env ERVWE1 placental expression, Journal of Virology 78: 12157-12168, 2004.
- Echert, D., Buhl, S., Weber, S., Jager, R., and Schorle, H., The AP-2 family of transcription factors, Genome Biology 6(13):246, 2005
- Zhao, C., and Meng, A., Sp1-like transcription factors are regulators of embryonic development in vertebrates, Development, Growth, & Differentiation 47(4):201-11, 2005.
- Yu, C., Shen, K., Lin, M., Chen, P., Lin, C., Chang, G., and Chen, H., GCMa regulates the syncytin-mediated trophoblast fusion, The Journal of Biological Chemistry 277: 50062-50068, 2002.
- Okulicz, W.C. and Ace, C.I., Temporal regulation of gene expression during the expected window of receptivity in the rhesus monkey endometrium, Biology of Reproduction 69:1593-1599, 2003.
- Dunlap, K.A., Palmarini, M., Varela, M., Burghardt, R.C., Hayashi, K., Farmer, J.K, and Spencer, T.E., Endogenous retroviruses regulate periimplantation placental growth and differentiation, Proceedings of the National Academy of Sciences, USA 103: 14390-14395, 2006.
- Caceres, M., NISC Comparative Sequencing Program, and Thomas, J.W., The gene of proviral origin syncytin-1 is specific to hominoids and is inactive in old world monkeys, The Journal of Heredity 97: 100-106, 2006.
- Dupressoir, A., Marceau, G., Vernochet, C., Benit, L., Kanellopoulos, C., Sapin, V., Heidmann, T., Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae, Proceedings of the National Academy of Sciences, USA 102: 725-730, 2005.
- Dunlap, K.A., Palmarini, M., Varela, M., Burghardt, R.C., Hayashi, K., Farmer, J.K, and Spencer, T.E., Endogenous retroviruses regulate periimplantation placental growth and differentiation, Proceedings of the National Academy of Sciences, USA 103: 14390-14395, 2006.
- Levine, K.L., Steiner, B., Johnson, K., Aronoff, R., Quinton, T.J., and Linial, M.L., Unusual features of integrated cDNAs generated by infection with genome-free retroviruses, Molecular and Cellular Biology 10: 1891-1900, 1990.
- Hajjar, A.M., and Linial, M.L., Characterization of a unique retroviral recombinant containing 7S L sequences, Journal of Virology 67: 7677-7679, 1993.
- Behe, M.J., Darwin’s Black Box, The Free Press, NY., p. 39, 1996.

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis

