The Coliform Kind: E. coli and Its “Cousins”
The Good, the Bad, and the Deadly
Abstract
Even though some intestinal bacteria strains are pathogenic and even deadly, most coliforms strains still show evidence of being one of God’s “very good” creations. In fact, bacteria serve an intrinsic role in the colon of the human body. These bacteria aid in the early development of the immune system and stimulate up to 80% of immune cells in adults. In addition, digestive enzymes, Vitamins K and B12, are produced by Escherichia coli and other coliforms. E. coli is the best-known bacteria that is classified as coliforms. The term “coliform” name was historically attributed due to the “Bacillus coli-like” forms. The bacteria are characteristically Gram-negative bacillus (rod-shaped) organisms found in the colons of man and animal. Typical genera include Escherichia, Citrobacter, Klebsiella, and Enterobacter. All these bacteria genera ferment lactose and are phenotypically similar.
These genera are often used as indicators in testing the quality of water. Their presence indicates contamination that can cause transmission of infection. This leads to a bad reputation for the coliform kind. How could God consider this creation good if they cause disease?
Introduction
Coliforms are just a few members of microbiota that live in our intestines. Coliforms are "coli-like" forms or species similar to E. coli. Literally, they are forms of bacteria from the colon, or other areas of the small or large intestine. In the human body, there are over 90 trillion cells belonging to over 1000 species by some estimates, and together they form a mass that has the metabolic activity of a “hidden organ” for the host. To give an idea of the size of the microbial community in the gut microbiome, the average human gut contains about two pounds of microbes (90 trillion individual microbes) that live in an intimate partnership with the human body! For comparison, the average human body is made of 10 trillion eukaryotic human cells. Some microbiologists estimate about 90 trillion bacteria; therefore only 1 in 10 cells in your body is human according to many microbiologists (though a recent study estimates 50%/50%).1 In spite of the amazing numbers, we are still distinct as humans, yet scientists are just now figuring how complex we are.
Gut Microbiota Numbers and Immunity
The further into the gastrointestinal tract, the more crowded the bacteria become. About 10 bacteria species per gram of contents are found in the esophagus, while about 100 bacteria species per gram reside in the colon (the large intestine). Their total amounts to 100 billion bacteria per gram egested in feces. About 40% of fecal matter weight is bacteria. This brings up a mysterious question: With 70% of the body’s immune response originating in the GI tract, how do commensal bacteria survive while the pathogens are destroyed? (Tortora, Funke, and Case 2018). Francis, Ingle, and Wood (2018) propose that bacteriophage (viruses that infect bacteria) “cloaking” of the bacterial cell surface as “an elegant mechanism whereby the normal microbiota bacteria are protected from immune detection and pathogenic bacteria can be directly lysed by the same phage. Additionally, both phage genome integration and cloaking can be used to prevent normal flora bacteria from conversion to a pathogenic state. Further support for the phage cloaking aspect of our theory has been demonstrated in recent studies which show that phage proteins bind specifically to microbial-associated molecular patterns (MAMPs), which are known to be the major ligands that activate the mammalian immune system.”
In the GI tract, the mucosal immunity involves the mucosal associated lymphoid tissue (MALT) the largest immune organ of the body. This includes Peyer’s patches that represent a major portion (Figs. 1 and 2). In a healthy individual, some estimate that 80% of immune cells are in MALT (Pommerville 2018). Mucosal immunity represents an interwoven network of tissues, cells, and molecules to protect against an infection in the intestine. Otherwise, pathogens could invade the epithelium if broken.
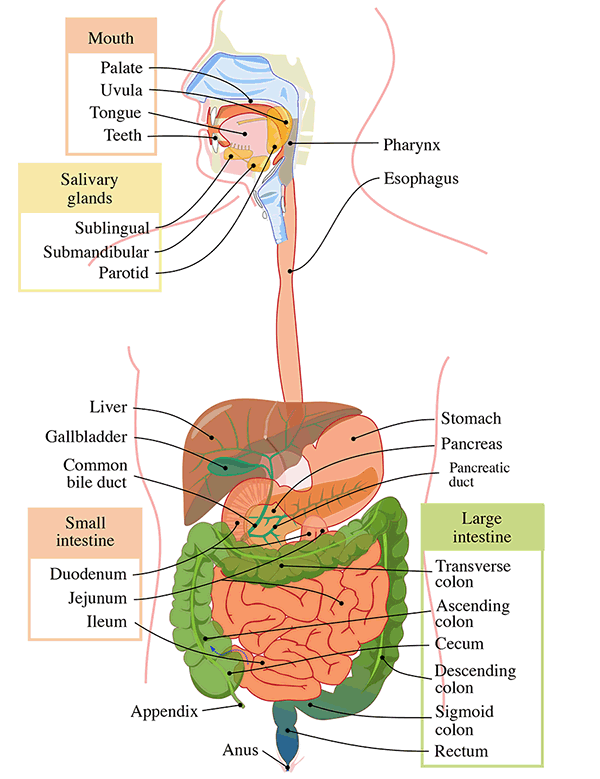
Figure 1. GI Tract Anatomy (Wiki commons). The gastrointestinal tract (digestive tract, gut, or alimentary canal) is a wonderfully made, designed organ system within humans and animals that takes in food, digests it to extract and absorb energy and nutrients, interfaces with the immune system, and expels the remaining waste as feces.
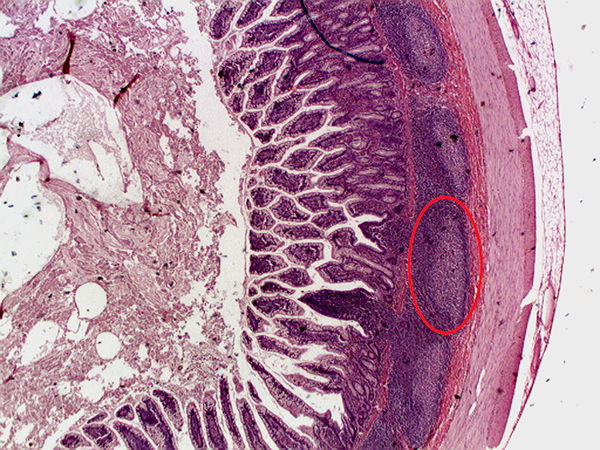
Figure 2. Peyer’s Patch in GI. (Wiki commons). A picture of Peyer's Patches, which are organized lymphoid nodules commonly found in the small intestines. This histology section was taken from the ileum, the final section of the small intestines.
Current research indicates resident microbes may have as much to do with sorting out good from bad “neighbors” and eliminating germs as the immune system does. Humans are born with very few microbes and an underdeveloped immune system. But, as we grow in bacteria, so does the immune system. The first bacteria in an infant “teach” the immune system to tolerate “good” bacteria. This is reinforced as beneficial compounds such as vitamins and other micronutrients, which are made by good coliforms. Other gram-negative bacteria, like Bacteroides, activate IgAs (Immunoglobulin A) in the intestinal mucosa fluid. These antibodies also have anti-inflammatory effects and prevent pathogens from attaching to epithelial cells. Furthermore, resident microbes also produce antimicrobial peptides (AMPs) that prevent germs from growing. Some bacteria induce our own AMPs and cause host cells to make other compounds, including cathelicidin. Cathelicidin-related antimicrobial peptides inhibit unwanted bacteria (Tortora, Funke, and Case, 2018).
Coliforms
Coliform bacteria are typically known as “indicator” microbes in surface water (pond, lake, stream, river) environments, since their presence may indicate the presence of pathogens or parasites in the water. In general, water quality monitoring agencies follow up a coliform-positive test with more specialized tests for serious pathogens or parasites (ex. cholera, Cryptosporidium, Giardia, etc.). It would be too expensive to routinely monitor for all serious disease-causing organisms. There are several criteria for being classified as indicator organism. The most important criteria is that the microbe must be consistently present in human feces in substantial numbers so that its detection is a good indicator of human waste entering water. The indicator should also survive in the water at least as well as the pathogens and must be detectable by simple tests that can be carried out by people with only basic training in microbiology. Coliforms are gram-negative bacilli that ferment lactose to a gas within 48 hours after incubation in 35°C (Table 1). Because some coliforms are not entirely enteric bacteria but can also be found in plant and solid samples, current standards distinguish fecal coliforms. E. coli is the best-known coliform (Fig. 3) and constitutes a large portion of the human intestinal population. However, in our own research studies, we found that total coliform counts were better indicators of Giardia and Cryptosporidium than E. coli alone.
Table 1 Coliform Facts
- bacteria in the family Enterobacteriaceae
- the most common genera Enterobacter, Klebsiella, Citrobacter, and Escherichia
- present in the intestinal tract of numerous organisms
- most are facultative anaerobes
- gram negative, non-spore forming rods
- ferment lactose with acid and gas production in 48 hours at 35°C
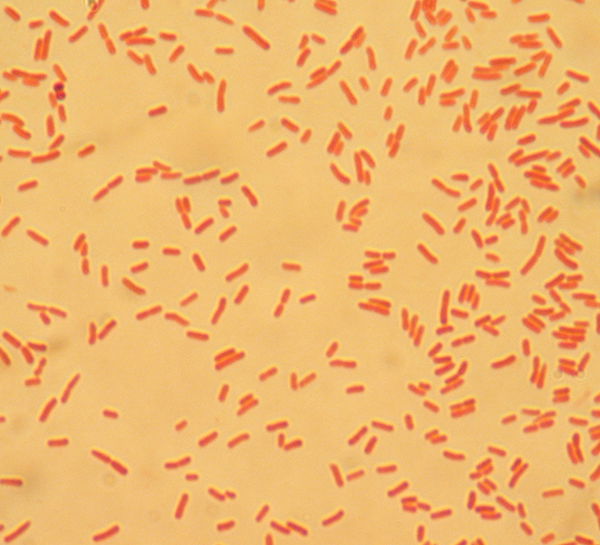
Figure 3. E. coli Gram stain (Wiki commons image). E. coli are Gram-negative bacteria, thus red or pink colored. The red color is due to a counterstain, called safranin.

Figure 4. Austrian pediatrician Theodor Escherich (Wiki commons image). Theodor Escherich (1857–1911) was a pediatrician and discovered E. coli in 1885.
In this paper, we describe only some of the most common types of coliforms. All coliforms are members in the Family Enterobaciaceae. E. coli does not use citrate as a source of carbon for “normal” metabolism, a distinguishing trait. Escherichia species can be motile with peritrichous flagella while others are non-motile. Most strains have pili (also called fimbriae) that extend to the bacterial surface to adhere to intestinal surfaces or biofilm environments. A basic list of coliform characteristics is contained in Table 1.
Escherichia coli, originally called “Bacterium coli commune,” was first isolated from the feces of a child in 1885 by the Austrian pediatrician Theodor Escherich (Fig.4) (Escherich 1885). Escherichia coli is a common inhabitant of the gastrointestinal tract of humans and animals. There are E. coli strains that are harmless commensals of the intestinal tract and others that are major pathogens of humans and animals.
Citrobacter (Fig. 5) is genus of flagellated bacteria that utilizes citrate as a source of carbon for normal metabolism. It moves by peritrichous flagella like many E. coli species. C. freundii is a common component of the gut microbiome of healthy humans (Schloissnig et al. 2013). While most strains are beneficial, there are significant phenotypic variations among strains, even those that share > 99% of their genome (Morowitz et al. 2011). The fermentation of lactose by these bacteria is delayed or absent (which is different from E. coli). They produce trimethlylene glycol from glycerol, a process first noticed by George Francis Gillen, a very distant cousin of the senior author (Fig. 6) (1930).
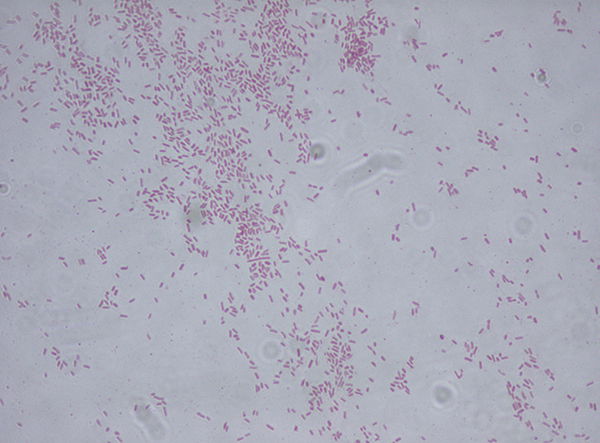
Figure 5. Citrobacter freundenii Gram stain (Wiki commons image). Citrobacter are Gram-negative bacteria, thus red or pink colored. The red color is due to a counterstain, called safranin.

Figure 6. Alan Gillen with his thesis and photo of George Francis Gillen.
Citrobacter as a genus was proposed in 1930 by George Francis Gillen2 in his master’s thesis at Iowa State under a different name. Then, two years later in 1932, bacteria producing trimethylene glycol was formally published by Chester H. Werkman and Gillen in the Journal of Bacteriology. Werkman was Gillen’s advisor at Iowa State, and they formally suggested the genus name as Citrobacter due to their ability to utilize citrate easily in normal metabolism. For many years, Werkman and his students for many years were interested in water sanitation and distinguishing various coliform bacteria. They were interested in both bacteria systematics and establishing practical guidelines for water safety: which bacteria were safe vs. pathogenic in water. Werkman also did a number of early studies (1920s) on vitamin production by E. coli. This trait and others would distinguish Citrobacter from E. coli, Enterobacter, and other coliforms known at the time.
Klebsiella (Fig. 7) is a bacterium often reported in the news due to one species that is a “superbug.” Klebsiella organisms are categorized microbiologically as gram-negative, facultative anaerobic, nonmotile bacteria. Klebsiella organisms reside in soil, water, and vegetation; some strains are considered a part of the normal microbiota of the human gastrointestinal tract. The genus is named for German physician and bacteriologist Edwin Klebs. The prototype species is Klebsiella pneumoniae. The first Klebsiella species ever described was a capsulated bacillus from patients with a chronic condition of the nose called rhinoscleroma.
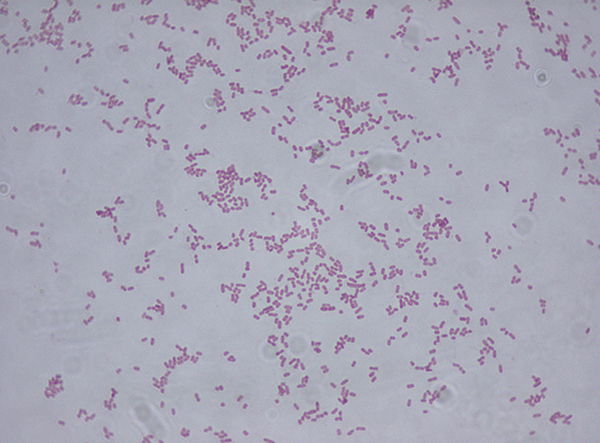
Figure 7. Klebsiella Gram stain (Wiki commons image). Klebsiella are Gram-negative bacteria, thus red or pink colored.
There are many other coliforms including those in the genus Enterobacter, Aerobacter, Hafnia and others; however, their presence in nature is beyond the scope of this article. These genera occur in humans and animals, including birds and mammals, but rarely cause infections.
The Coliform “Kind”3 and Good Design
Humans cannot properly digest food without bacteria. Coliforms are a normal inhabitant of the large intestine of vertebrates, including humans, and their presence is beneficial as it helps to produce certain vitamins and breaks down otherwise indigestible foodstuffs. E. coli are part of our normal microflora (also called normal flora) in human intestines (Fig. 1). Vitamin B12 is formed in some fermented Indonesian foods by particular strains of Citrobacter freundii and Klebsiella pneumoniae (Keuth and Bisping,1994) and provides nutritional benefits for those in developing countries. Both Citrobacter freundii and Klebsiella pneumoniae also play a role in the nitrogen cycle in the environment.
Coliforms and other enteric bacteria provide vitamin K, vitamin B12, thiamin, and riboflavin. In addition, they take up space and keep out pathogens and parasites. If one takes long-term antibiotics, they can wipe out these normal good microbiota, and diarrhea can result in addition to many other medical implications. The coliforms and other beneficial enteric bacteria protect us against colonization of those microbes or parasites that can cause diarrhea and even worse symptoms. Coliforms stimulate antibody production within one to two days after birth. Theodor Escherich (1885) discovered this in his pediatric stool studies in the 1800s when he first described E. coli and other related coliforms. They also stimulate immune system development. In turn, the man-microbe interface can prevent colon cancer, autoimmune diseases, and more. The intimate microbiome is an interwoven good design of the body.
The Interwoven Complexity of the Gut Microbiome
An interwoven adaptation package is a biological relationship in which the whole is functionally more than the sum of its individual components. We call this an extension of the interwoven complexity observed in the human body (Gillen 2006). Dr. Joseph Francis (Francis 2009) calls it the “biomatrix” of life, or “organosubstrate” theory. In more recent years, design advocates have used terms like immune system as a microbial interface system. This creation model states that the immune system was designed to interact with microbes like coliform bacteria, gut protozoans, and fungi.
Indeed, the intestinal bacteria contribute to the general well-being of both microbes and people by synthesizing a number of the vitamins essential for good nutrition and breaking down various macronutrients. The human body cannot synthesize niacin to make nicotinamide adenine dinucleotide (NAD) that is necessary for energy conversion in the cell’s mitochondria. The B vitamins, including niacin, cobalamin (B12) biotin, thiamine, and riboflavin are necessary for normal energy levels, freedom from fatigue, and proper functioning of nerves. A prolonged deficiency in any one of these vitamins may lead to chronic fatigue and inability to lead a normal life. As for the benefit to coliforms (and other enteric bacteria), the human colon provides them a stable nutritional home. In short, both parties benefit from this mutualistic relationship (Table 2).
| Vitamins and Immunity | Physiological Role | Disease if Deficient |
|---|---|---|
| 1. Vitamin K | Needed in blood clotting | Hemorrhage in newborns who lack gut bacteria |
| 2. Vitamin B12 (absorbs) | Coenzyme needed in protein formation | Pernicious anemia; lack of energy |
| 3. Niacin (Vitamin B3) | Part of coenzymes in energy metabolism | Pellagra; lack of energy; fatigue |
| 4. Vitamin B-Complex (thiamine, biotin, and riboflavin) | Carbohydrate metabolism Coenzymes in cell respiration |
Beriberi, loss of appetite, fatigue, inflammation and breakdown of skin |
| 5. Immune cells IgA, AMPs, Dendritic cells | Development of the immune system | Poor immune response to pathogens and parasites |
Pre-Fall World, Probiotics, and the Gut Microbiome
Where do the E. coli microbes fit into God’s “very good” creation? Only about 1% of all bacteria are pathogenic.4 Many live in a mutualistic relationship with other organisms. Mutualism is a type of symbiosis (Greek sym, together; bios, life) where both sides benefit. God’s very good creation included these important microbes.
The gut microbiome provides clues to the pre-Fall function of bacteria (Fig. 8). Dr. Francis (Francis 2009) suggested that microbes are a natural and healthy extension of the body and that bacteria form a biomatrix. When God created man, most likely he made the Escherichia/Citrobacter kind to help maintain the digestive and immune system. Perhaps, the original Escherichia kind diversified by microhabitat: some for the small intestine (ileum, duodenum, and jejunum), and some in the ascending, transverse, descending colon, and rectum.5
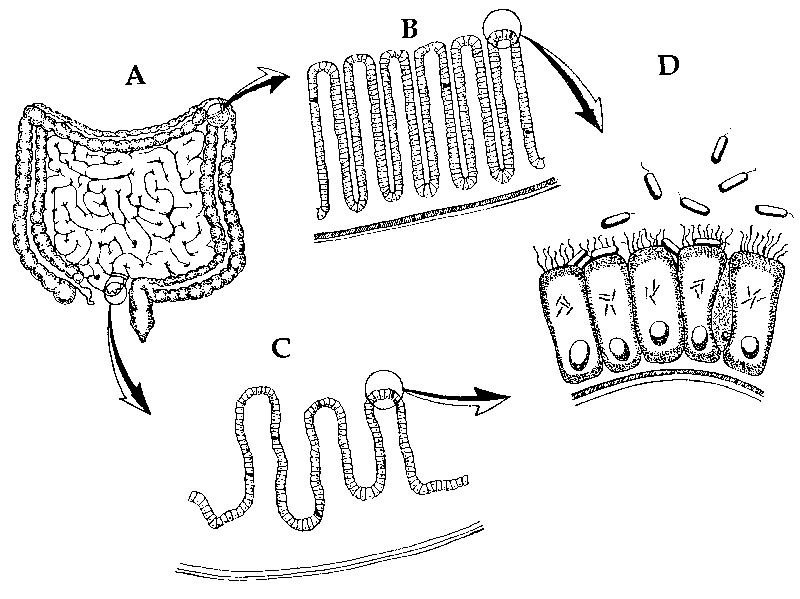
Figure 8. Coliform bacteria in the intestines. Most of the time, bacteria inhabit the gastrointestinal (GI) tract mutualistically. A illustrates the small and large intestines. B is a close up of the villus lining. C is a close up of the microvillus lining, the site where nutrients are absorbed and transferred to the bloodstream. D illustrates the site where coliforms assists in the absorption of micronutrients and manufactures B-vitamins at the surface of the villi (adapted from Gillen, Sherwin and Knowles 2001, p. 107; Gillen, 2009).
Human infants are born nearly sterile and acquire microbial inhabitants with age. The first 100 days are the most important in forming this biome. Many diseases result from imbalances during this stage. One such manner of interruption is a cesarean birth because the microbes are not gained from the birth canal. At early stages of growth, the microbiome is similar at the mouth, gut, and skin. The forms differentiate with age. Citrobacter species are a main player in this biome. Citrobacter koseri is an example of bacteria that grows only in the gut and helps to differentiate the areas (Ohm 2010). Citrobactor spp. imbalance has been linked to food allergy (Savage 2017). An inverse relationship has been found between the presence of Citrobacter and allergic sensitization. This is one more example of the importance of the human microbiota early in life.
The Bad
Escherichia coli and various Citrobacter species in the digestive tract of man and animals can be good. However, when they are displaced to the urinary tract (urethra, ureters, and kidneys), they can cause urinary tract infections. Urinary tract infections can vary from slight irritations to symptoms that are severe. E. coli is the culprit of the majority of urinary tract infections. I have written on E. coli, and some other effects on the body have been previously reported (Gillen 2007; Gillen and Oliver 2010). In this paper, we report a current trend of more UTIs caused by Citrobacter and also the groundwork by George Francis Gillen. The author reports the emergence of Citrobacter as a common urinary pathogen in hospitalized patients. The genus Citrobacter was discovered in 1932 by Werkman and Gillen. These organisms are found naturally in soil, water, intestinal tract of animals, and now increasingly in human clinical samples, especially UTIs.
Hospital-acquired urinary tract infection (UTI) is the most common healthcare-associated infection (nosocomial infection), accounting for 13% of the total healthcare infections, thus posing a serious health threat. Additionally, the prevalence of antimicrobial resistance among urinary pathogens has increased worldwide due to uncontrolled and indiscriminate antibiotic usage. Furthermore, certain pathogens that were isolated sporadically have now emerged as prominent healthcare-associated pathogens.
Citrobacter gillenii
Citrobacter gilleni was first described in the 1990s to honor George Francis Gillen. Citrobacter gilleni was formerly called Citrobacter genomospecies 10. It is distinguished for a few traits, such as delayed positive color change/growth on citrate medium. Little is known about this species other than disease due its rarity and fairly recent discovery. It is primarily known for its being a nosocomial infection and, in particular, its increasing numbers in Europe as a urinary tract infection.
The Deadly
Some coliform bacteria in high numbers are not only indicators of bad water quality for other bad and deadly diseases, like giardiasis, cryptosporidiosis, and cholera, but also can be deadly themselves. We will discuss only two that are very common: E. coli O157:H7 and Klebsiella pneumoniae.
E. coli O157:H7
E. coli O157:H7 is a “lion” of a bacterium (Fig. 9). It has the deadly ability to inject toxins into our cells (specifically kidney and intestinal cells), cause internal bleeding and shut down vital organs. This usually results in hemolytic uremic syndrome and a critical care situation in a hospital emergency room. This strain was first recognized in 1982, and since then has emerged as a public health problem. It is now one of the leading causes of diarrhea worldwide. In 1996, some 9,000 people in Japan became ill and seven died. The recent outbreaks of E. coli O157:H7 in the U.S., associated with contamination of spinach, undercooked meat, and unpasteurized beverages (like apple juice), have alerted public health officials that new methods of testing for bacteria in food must be developed. For decades now, there have been new outbreaks in the nation every year (Gillen and Oliver 2010; Tortora, Funke, and Case 2018).
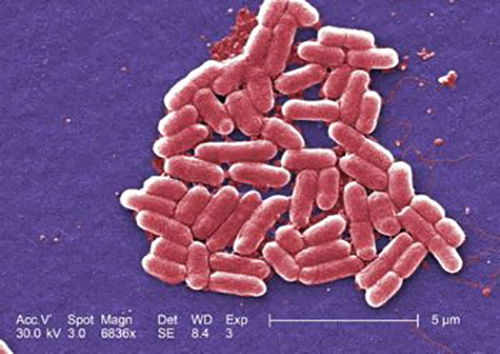
Figure 9. E. coli O157:H7 is a “lion” of a bacterium (CDC image # 10068) Virulence factors include fimbriae, adhesions, type III secretion system; and produce Shiga-like toxin.
Klebsiella pneumoniae
Klebsiella pneumoniae in the environment appears to play a positive role in nitrogen fixation in plants like wheat. Other species of Klebsiella fix nitrogen in other plants in the environment and clearly play a positive role in nature. While K. pneumoniae is a bacterium that normally inhabits the gut tracts of man and animals, it can cause pneumonia and sepsis when displaced in the lungs. Klebsiella pneumoniae (Fig. 10) is a hospital acquired pathogen (HAP). It is a gram-negative rod with a prominent capsule. It makes up to 15% of HAP in the USA and is often acquired by respiratory droplets; however, it can sometimes naturally be acquired in the nasopharynx. As a primary pneumonia, it is characterized by sudden onset and a gelatinous, reddish-brown sputum. The color comes from blood seeping into alveolar sacs of the lungs as the bacteria multiply. This tissue damage can lead to death. In its secondary form, it occurs in immunocompromised people and spreads via intravenous routes from infected clothing, foods, and hands of unskilled healthcare workers in hospitals, clinics, assisted living facilities, and nursing homes. It is becoming an increasingly serious and fatal pathogen.

Figure 10. Klebsiella pneumoniae (CDC image #16809). Centers for Disease Control (CDC) laboratorian Alicia Shams, as she was showing viewers a Petri dish culture plate that demonstrated growth of Klebsiella pneumoniae on a growth medium of MacConkey agar. Using an inoculating loop held in her right hand, Ms. Shams was pointing out the bacterial colonies which grew atop the MacConkey medium contained in the Petri dish she held in her left hand. In healthcare settings, Klebsiella infections commonly occur among sick patients who are receiving treatment for other conditions. Patients whose care requires devices like ventilators (breathing machines) or intravenous (vein) catheters, and patients who are taking long courses of certain antibiotics are most at risk for Klebsiella infections. Healthy people usually do not get Klebsiella infections.
Some have called this a “superbug,” a nightmarish bacteria due its resistance to every known antibiotic in some strains. Klebsiella has routinely been treated with carbapenems, but strains of carbapenem-resistant Enterobacteriaceae (CRE) have now been detected in the US. Although MRSA is a more common and frightening problem,6 carbapenem-resistant Klebsiella, though much less common, is far more dangerous. While there are still a number of antibiotics that can kill MRSA, Klebsiella is a much tougher cookie. There are few good alternatives for treatment; however, they seldom work today in light of its high antibiotic resistance.
Water Quality Studies: Indicators of Pathogens and Parasites
Coliform bacteria are not innately pathogenic (disease causing) organisms; they are only mildly infectious and relatively easy to culture. For this reason, these bacteria are relatively safe to work with in the laboratory as indicator organisms. The government standard requires public drinking water supplies to demonstrate the absence of total coliform per 100 mL (about 4 oz.) of drinking water.
Local Research on Coliforms
In our own research, the efficacy of using indicators such as total coliforms and single enteric bacteria species to predict the presence of protozoan parasites such as Giardia and Cryptosporidium was examined in the surface water of a small lake. Parasite and bacterial presence was measured over a nine-month period in two designated zones. Enteric bacteria including Escherichia coli, Citrobacter freundii, Citrobacter amalonaticus, and Klebsiella oxytoca were detected in the sampled water (Fig. 11). Along with bacterial indicators, weather, biofilm, turbidity, and wildlife prevalence played a significant role in predicting the protozoan parasite concentrations. The presence of Giardia was indicated by Klebsiella oxytoca, Enterobacter cloacae, and Citrobacter amalonaticus, all of which are duck coliforms. A trend was observed indicating that as total coliform levels and turbidity increased, so did the levels of Cryptosporidium. None of the data collected indicated that a sole indicator organism was an adept predictor of Cryptosporidium presence. Thus, the presence of multiple indicator organisms (C. freundii, total coliforms, biofilm) is a more suitable predictor for Cryptosporidium. The exact source of the Cryptosporidium remains unknown. For the benefit of public health, the observation of the total collection of indicators (Fig. 12) should be implemented during water quality testing. More study is needed to both identify the parasite source and to further illuminate the specific microbial indicators of surface waters at Candler Mountain and its associated Library Lake at Liberty University, as well as the nearby James River.
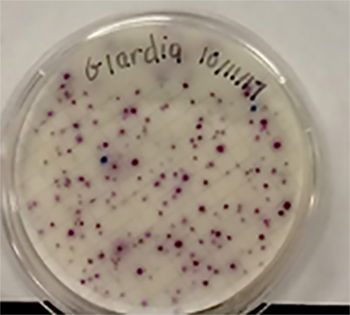
Figure 11. Coliforms isolated from LU Library Lake Giardia zone. In the MF Coliscan method, E. coli will produce both galactosidase and glucuronidase and will therefore grow as dark blue to purple colonies in the medium. It is simple to count the blue/purple colonies (E. coli) which indicate the number of E. coli per sample. The pink colonies indicate the number of general coliforms per sample. The combined general coliform and E. coli number equals the total coliform number.
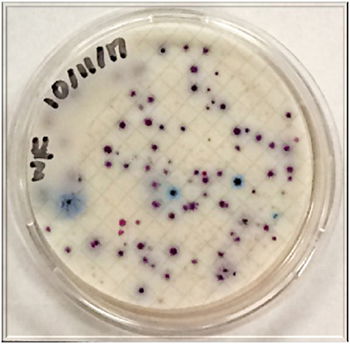
Figure 12. Coliforms isolated from LU Library Waterfall zone. In the MF Coliscan method, E. coli will produce both galactosidase and glucuronidase and will therefore grow as dark blue to purple colonies in the medium. It is simple to count the blue/purple colonies (E. coli) which indicate the number of E. coli per sample. The pink colonies indicate the number of general coliforms per sample. The combined general coliform and E. coli number equals the total coliform number.
Citrobacter—Another "Kind" of E. coli
Citrobacter freundii was originally described as Escherichia spp. They are 92% similar at the genome level. Citrobacter and E. coli are very similar in size, shape, and traits, except they differ in the way they utilize citrate and in other metabolic features. If we define species in the way the newest Bergey’s Manual of Systematic Bacteriology (Brenner 2005) defines it, then these two are distinct genera and distinct species. From a creation view, they are likely the same baramin (kind) and all descendants of the same original “coliform.” Bergey’s manual has been the leading reference for identifying and classifying bacteria since the 1920s.
Disease Prevention in the Bible
The key to clean, potable drinking water control is absence of pathogens, parasites, and chemicals. But for a Christian, seeking to be like Jehovah Rapha, the Great Physician, is important. God has revealed himself as Jehovah Rapha in the context of bitter (non-potable) water occurring in the wilderness:
So Moses brought Israel from the Red Sea; then they went out into the Wilderness of Shur. And they went three days in the wilderness and found no water. Now when they came to Marah, they could not drink the waters of Marah, for they were bitter. Therefore, the name of it was called Marah. And the people complained against Moses, saying, “What shall we drink?” So he cried out to the Lord, and the Lord showed him a tree. When he cast it into the waters, the waters were made sweet.
There He made a statute and an ordinance for them, and there He tested them, and said, “If you diligently heed the voice of the Lord your God and do what is right in His sight, give ear to His commandments and keep all His statutes, I will put none of the diseases on you which I have brought on the Egyptians. For I am the Lord who heals you.” (Ex. 15: 23–26)
Rapha means “to cure, to repair, to heal, to mend back together,” or “to restore one to health or usefulness” (Gillen 2006). Usually the word heal or healing is used in conjunction with Jehovah Rapha. Healing implies that God is involved in restoration. This is the nature of Jehovah Rapha. Indeed, the Great Physician prevents diseases by warning us of dangers in contaminated waters. With similar warnings, the Jews were instructed to contain human waste. No one knows with certainty the origins of E. coli diarrheal disease. However, it appears that it is a very old disease. The Lord gave instructions to the Israelites to dispose of their human waste outside the camp for a sanitary system that is not dissimilar to what we use today:
Also you shall have a place outside the camp, where you may go out; and you shall have an implement among your equipment, and when you sit down outside, you shall dig with it and turn and cover your refuse. (Deuteronomy 23:12–136)
Today, in the Western world, this can be practically applied by using toilets and a septic tank or sewage treatment systems to remove potentially infectious human waste from living areas. Throughout history, mankind has not always heeded this directive given through Moses and has suffered the consequences of severe diarrhea, dysentery, and other gastrointestinal maladies including severe dehydration, which could even lead to death.
The Good, Bad, and Deadly Summary
There is great evidence that enteric bacteria were created for good purpose. We have documented dozens of benefits for the gut microbiome, and no doubt biologists will find dozens more in the decades to come. However, over time, the curse, corruption, and contagion followed the once good creation, and the fall, decay, and death occurred. Some of the very good microbes were modified, displaced, and overgrew their boundaries—some bad and disruptive, others deadly. The genesis of new germs seems to take place annually (Gillen 2007). Only in the new heaven and earth will there be an exodus of pathogenic organisms and a total restoration of the original creation.
References
Brenner, D. J., et al. 2005. Bergey's Manual® of Systematic Bacteriology: Volume 2: The Proteobacteria.
Escherich T. 1885. “The Intestinal Bacteria of the Neonate and Breast-fed Infant.” Rev Infect Dis., 1988, vol. 10, pp. 1220–25. (Note: This is the English Translation in 1988).
Francis, J. 2009. “What about Bacteria?” Edited by Ham, K. (editor). The New Answers Book 3. Green Forest, Arkansas: Master Books.
Francis, J.W., et al. 2018. “Bacteriophages as Beneficial Regulators of the Mammalian Microbiome.” In Proceedings of the Eighth International Conference on Creationism, ed. J.H. Whitmore, pp. 152–157. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Gillen, A. L., et al. 2001. The Human Body: An Intelligent Design, 2nd ed. St. Joseph, Missouri: Creation Research Society Books.
Gillen, A. L. 2006. Body by Design: Fearfully & Wonderfully Made. 5th printing. Green Forest, Arkansas: Master Books.
Gillen, A. L. 2007. The Genesis of Germs: Disease and the Coming Plagues in a Fallen World. Green Forest, Arkansas: Master Books.
Gillen, G. F. 1930. “Bacteria Producing Trimethylene Glycol.” Master’s thesis. Iowa State College.
Gillen, A. L. and Oliver, J. D. 2010. “The Genesis of Pathogenic E. coli.” Answers In-Depth. October 6, 2010. https://answersingenesis.org/biology/microbiology/the-genesis-of-pathogenic-e-coli/
Morowitz, Michael J. et al. 2011. "Strain-resolved community genomic analysis of gut microbial colonization in a premature infant". Proceedings of the National Academy of Sciences. 108 (3): 1128–1133. doi:10.1073/pnas.1010992108. ISSN 0027-8424. PMC 3024690. PMID 21191099.
Olm, M., et al. 2018. “Identical Bacterial Populations Colonize Premature Infant Gut, Skin, and Oral Microbiomes and Exhibit Different in Situ Growth Rates.” Genome Research. Cold Springs Harbor Laboratory Press. https://genome.cshlp.org/content/27/4/601.full.pdf+html
Pommerville, J. 2017. Fundamentals of Microbiology. 11 ed. Burlington, MA: Jones & Bartlett Learning.
Savage, J. H., et al. 2017. “A Prospective Microbiome Wide Association Study of Food Sensitization and Food Allergy in Early Childhood. Wiley Online Library. https://onlinelibrary-wiley-com.ezproxy.liberty.edu/doi/abs/10.1111/all.13232
Schloissnig, Siegfried, et al. (2013). "Genomic variation landscape of the human gut microbiome". Nature. 493 (7430): 45–50. doi:10.1038/nature11711. ISSN 0028-0836. PMC 3536929. PMID 23222524.
Tortora, G. J., et al. 2018. Microbiology, an Introduction. 13th ed. San Francisco, California: Pearson Co.
Werkman, C.H. and Gillen, C. F. 1932. Bacteria Producing Trimethylene Glycol. J. Bacteriol. February 1932. Vol. 23 no. 2, 167–182.
Footnotes
- The recent update (August 2018) to the popular Tortora, Funke, and Case (2018) microbiology text states:
"An adult human body is composed of about 30 trillion body cells and harbors 40 trillion bacteria cells." This actually differs from their previous printed versions of the same text. Biologists continue to study and refine cell counts as new technologies are available. See also https://www.biorxiv.org/content/early/2016/01/06/036103 George Francis Gillen was Episcopalian Christian. The surname, Gillen means servant of St. John. Historically, most “early” Gillens were members of the Anglican church in northern United Kingdom and Ireland.
- Bodie Hodge, "A Biblically Based Taxonomy?" Answers in Genesis (June 25, 2010), https://answersingenesis.org/creation-science/baraminology/a-biblically-based-taxonomy/
- MedlinePlus, (2018), s.v. "Bacterial Infections," https://medlineplus.gov/bacterialinfections.html
- Alan L. Gillen and Douglas Oliver, "The Genesis of Pathogenic E. Coli," Answers in Depth (October 6, 2010), https://answersingenesis.org/biology/microbiology/the-genesis-of-pathogenic-e-coli/
- Alan L. Gillan and Rachel Walters, "Super Staph in the Community: Is It Evolving?" Answers in Depth (October 22, 2015), https://answersingenesis.org/biology/disease/staph-is-it-evolving/
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis


