Bacteria’s Unique Design—Pooling Resources
Semi-Technical
God created bacteria to perform vital roles in every imaginable environment. To help them perform those roles, He gave them unique designs to share information so they’re prepared for new challenges.
It’s a dark and stormy night, and the bacteria living in a small puddle outside the factory are struggling to survive. Nutrients are scarce, and the man-made substances found in the wastewater are unusable by the bacteria. Will they perish? The answer will surprise you!
Unlike humans and many animals, bacteria typically do not have the same choices in dealing with their environment. If they live outside and it’s raining, they can’t put up an umbrella or go inside. If nutrients are low, they can’t go to the grocery store and get food. Instead God designed bacteria to be “master adapters,” with genetic resources that humans and animals don’t have.
An Ocean of Options
Originally God designed bacteria to be invaluable servants to plants and animals in His “very good” world (Genesis 1:31); and the vast majority of bacteria still display beneficial characteristics. In the human gut, for instance, they break down nutrients and produce vitamins that we can’t make on our own and we couldn’t survive without. They also make many essential elements, such as sulfur, phosphorus, and carbon—from soil, water, and air—available to living things. In our fallen world, bacteria even break down deadly pollutants, such as waste products from factories.
God in His wisdom and grace designed these microscopic marvels with special genetic tools to adapt and survive in a fallen world, so that they can continue these servant roles. Bacteria are very small, and there is no way they could carry the full complement of genes they might need for every environmental situation they would face. It would also waste time and energy to replicate all that DNA every time the bacteria reproduced. Instead the full complement of genes, called the pan-genome, is “split” among varying strains of the same species. Furthermore, some species are capable of acquiring genes from other bacterial species.
As a result of genetic exchange among bacteria, the genome for any given bacterial genus or species is not easily defined. The material available to bacteria is like a “genetic ocean” that is constantly ebbing and flowing through bacteria, often via viruses (called bacteriophages) that infect bacteria and acquire and carry bacterial DNA to other bacteria. An estimated 1031 bacteriophages (ten thousand billion billion billion) fill the earth, and bacteriophages infect 1024 bacteria per second.1 There is a tremendous amount of genetic material flowing in and out of bacteria at any given time!
For just about any species other than bacteria, if you sequence the genome (the DNA present in the nucleus) of one member of the species, it will be very similar to every other member of that species. For example, my DNA is only 0.1% different from that of each person reading this article. This similarity allows scientists to talk about the human genome as opposed to human genomes.
But for just about any bacterial species, if you sequence the genome of one strain, it will likely be very different from other strains within that same species. In fact if you sequence 20 different strains, you will find many genes unique to only one strain! Again, this is what scientists often refer to as a pan-genome since it is present across many strains in a given species.
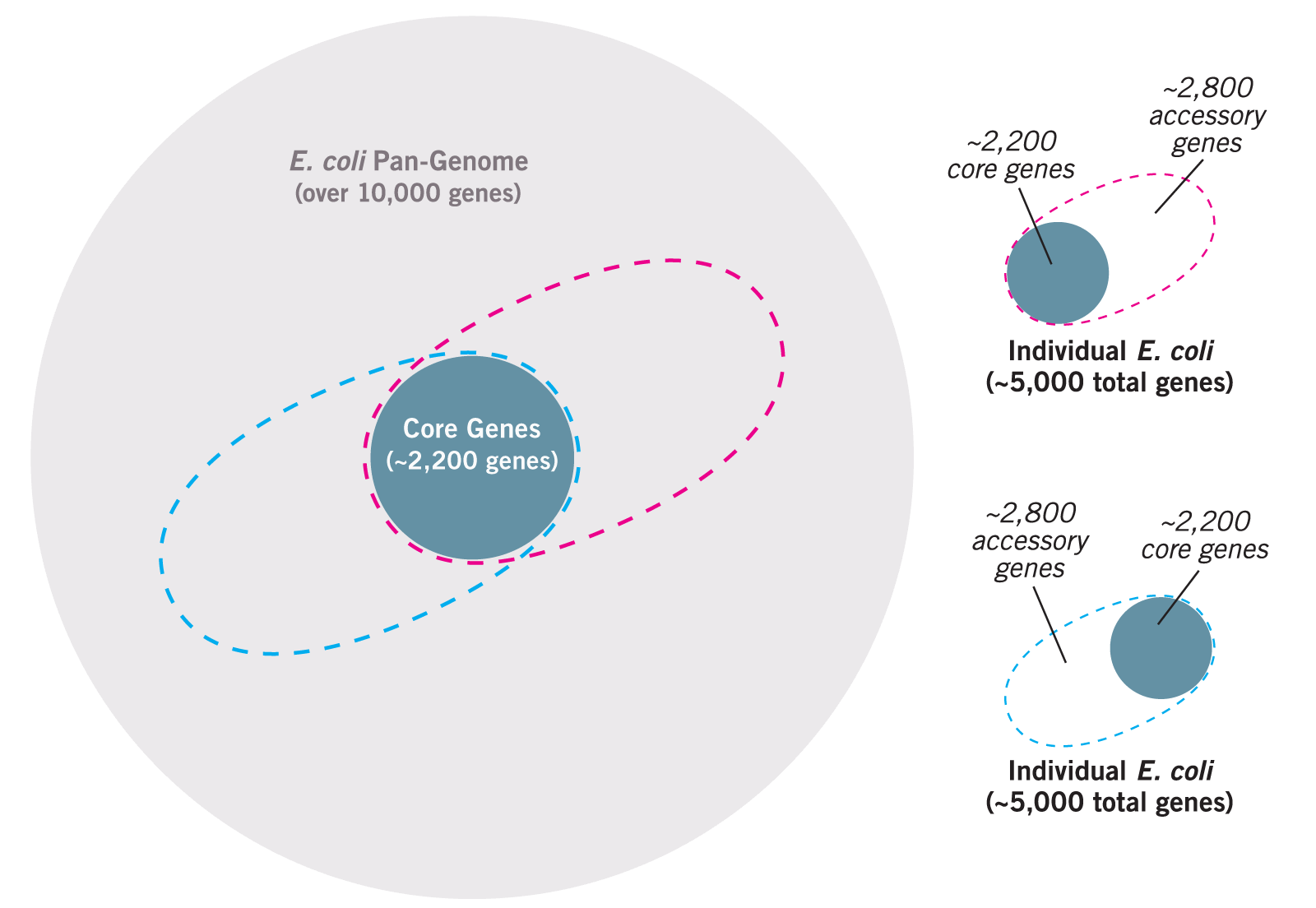
Consider the example of Escherichia coli, a common gut bacterium. The genome of each strain consists of approximately 5,000 genes. About 2,200 of those genes are found in virtually every other strain of E. coli sequenced.2 These genes (and the proteins produced from them), called core genes, are considered housekeeping genes and are needed by every strain for basic cellular functions. The remaining 2,800 genes are specific to that strain and called accessory or adaptive genes. These genes help it adapt and function in a specific environment.3 For example, a study that compared proteins produced by two harmful strains and one harmless strain of E. coli found that nearly 40% of the proteins produced by all three strains were the same, but 47% of the proteins were produced by only one strain.4
Think about the potential size of the E. coli pan-genome (all accessory genes in all strains). It is currently believed to be 10,000–13,000 genes and growing. For every new strain of E. coli sequenced, in fact, researchers expect (based on mathematical extrapolations) that 300 new genes will be discovered!5 It is very possible that the pan-genome of some bacterial species could consist of tens of thousands of genes, with some pangenomes being larger than the human genome of only 28,000–30,000 genes.
Tools of Adaptation
How do bacteria acquire this great diversity of genes? Two major mechanisms are available to help bacteria adapt to changing environments: mutations (in conjunction with natural selection) and horizontal gene transfer (HGT). Each mechanism is marvelously controlled to allow maximum adaptability within God’s specific goals for each species.
Mutations (and Natural Selection). Mutations are a change in the DNA sequence. Since DNA contains the instructions for making proteins, mutations can alter the structure or function of proteins. Mutant proteins can accomplish different tasks that allow the bacteria to survive better in a given situation (for example, break down a man-made substance in wastewater from a factory and use it for nutrition). The better-equipped bacteria will survive via natural selection and grow, reproduce, and eventually become the dominant bacteria.
For example, in the mid-1970s, bacteria were discovered in ponds with wastewater from a nylon factory. These bacteria (Arthobacter sp. K172) could digest the byproducts from manufacturing nylon, a man-made polymer that had never existed before it was produced in the 1940s. Thus, these bacteria must have acquired their ability to break down nylon in only a few decades.
How could they do this? In 2007, genetic analyses showed that mutations in a pre-existing gene in the bacteria, known as EII, produced a protein capable of breaking down nylon.6 The normal version of EII was found to break down a substance very similar to nylon. The mutations were essentially “tweaks” that allowed the bacteria to break down nylon and adapt and survive in a seemingly inhospitable environment.
Horizontal Gene Transfer. Another way that bacteria adapt is to borrow genetic material from other bacteria by a process known as horizontal gene transfer. This is different from vertical gene transfer, which occurs with traditional reproduction from parent to child. Instead, bacteria pass along their genes to other, separate bacteria.
There are three main ways this amazing transfer is accomplished: transformation, conjugation, and transduction. In transformation, the bacteria take in free-floating DNA directly from the environment. When bacteria die, the cells rupture and their DNA becomes part of the environment. Nearby living bacterial cells can then take up the DNA, which becomes part of their own genetic makeup.
Despite its wonderful flexibility, bacterial adaptation doesn’t change a bacterium into a completely different kind of organism.
Conjugation is the transfer of genetic material from one bacterium to another through a physical connection, a “bridge,” between the two. It’s the bacterial equivalent of sexual reproduction.
Transduction is the transfer of genetic material from one bacterium to another by viruses (the bacteriophages mentioned above, and yes, even bacteria can have viral infections!). The viruses are able to incorporate bacterial DNA into their own viral DNA and transfer it into the next bacteria they infect.
Horizontal gene transfer is nearly exclusive to bacteria because they need to adapt to such a wide variety of different environmental situations to survive. For example, antibiotic resistance genes and virulence (disease-causing) genes are often transferred among bacteria.
Unfortunately, in our fallen world, mutation and horizontal gene transfer don’t work perfectly. Sometimes in the process of adapting to a new environment, bacteria acquire genetic changes that also lead to disease. Understanding why some changes are bad is an active area of creation science research.
Anything New?
Despite its wonderful flexibility, bacterial adaptation doesn’t change a bacterium into a completely different kind of organism because it doesn’t result in the origin of novel genetic information needed for this type of change. In the case of bacteria, it is the alteration or exchange of pre-existing genetic information.
Mutations and natural selection have never been observed to originate novel information of the type necessary for molecules-to-man evolution. This is consistent with Scripture, which states that God created organisms according to their kind, implying that they were to reproduce according to their kind (Genesis 1, 6, and 8). We observe mechanisms that allow bacteria to adapt within limits but not to evolve into different kinds of organisms.
Just like with mutation and natural selection, horizontal gene transfer does not result in the origin of novel genetic information necessary for molecules-to-man evolution. The genetic information was pre-existing in other bacteria and was merely transferred from one bacterium to another. It’s like having money in your right pocket and putting it in your left pocket. You haven’t become any richer; you’ve just changed the location of your money.
Conclusion
God designed bacteria with these elaborate mechanisms so they could adapt quickly and share their genes in a post-Fall, post-Flood world. This has allowed them to refill the earth (Genesis 8:16–19) and continue their many good functions. The availability of a wide variety of genetic material that can be acquired quickly is absolutely essential to the bacteria’s master adapter abilities.
We’re still in the early stages of understanding this amazing design. So far, pan-genome studies have concentrated on comparing harmful strains to harmless ones to determine disease-causing genes and create better treatments. But more wonders are in store, as the Creator’s wise designs come to light.
Although nearly 40,000 bacterial genomes have been sequenced to date, this represents only a very small fraction of the actual number of bacterial species thought to exist (in the hundreds of thousands to millions). Many species cannot be cultured (grown) in the lab, making study difficult. In addition, only a handful of the trillions of bacteriophages thought to exist have been sequenced. To put it plainly, a lot of information is floating out there in the “genetic ocean” that we know nothing about! As better sequencing and culturing technologies are developed, it will be exciting to see in even greater depth God’s grand design for bacteria, His master adapters.
Answers Magazine
January – March 2015
This issue clears up ten common misconceptions about the Flood. Also discover the most profound evidence for creation on the planet.
Browse IssueFootnotes
- Hervé Tettelin, et al., “Genome Analysis of Multiple Pathogenic Isolates of Streptococcus agalactiae: Implications for the Microbial ‘Pan-genome,’” Proceedings of the National Academy of Sciences (2005) 102:13,950–13,955.
- Marie Touchon, et al., “Organised Genome Dynamics in the Escherichia coli Species Results in Highly Diverse Adaptive Paths,” PLOS Genetics (2009) 5:e1000344.
- Ibid.
- R. A. Welch, et al., “Extensive Mosaic Structure Revealed by the Complete Genome Sequence of Uropathogenic Escherichia coli,” Proceedings of the National Academy of Sciences (2002) 99:17,020–17,024.
- Alex Mira, et al., “The Bacterial Pan-genome: A New Paradigm in Microbiology,” International Microbiology (2010) 13:45–57.
- Kevin Anderson and Georgia Purdom, “A Creationist Perspective of Beneficial Mutations in Bacteria,” Proceedings of the Sixth International Conference on Creationism (Pittsburgh: Creation Science Fellowship), 73–86.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis