Beyond Darwin's Tree: A Critical Look at Phylogenetics
Abstract
The history of phylogenetics and its uses has been quite contentious. Many illogical arguments have been presented alongside a few more sensible ones. It is helpful to have an idea about what is presented, both good and bad, so that each can be logically refuted. Thus, this article contains a sampling of a wide range of arguments that evolutionists have used. The overarching idea, however, is that evolution predicts a nested hierarchy of life, with extant taxa continuous or discontinuous depending on their relationships. Because phylogenies are performed under the assumption that similarity is equivalent with ancestry, similarity and ancestry are interpreted accordingly. The methodology itself can be used by creationists, as long as all similarity is not assumed to be the result of ancestry but could rather the possible result of a common Designer.
Introduction
If you have ever opened a secular textbook, you have seen an example of phylogenetics, though it may not have been called that. Phylogeny is “the evolutionary history of a species or group of species,”1 and phylogenetics refers to the methods used to determine that history. Using features of living organisms referred to as “characters,” taxonomists attempt to reconstruct the evolutionary history of a group in what is called phylogenetic trees. The combination of phylogenetic trees of all organisms forms the Darwinian tree of life, which purports to represent the life history of all organisms. However, phylogenetics is undermined by some massive assumptions that blunt its force as an argument against creation.
History of Phylogeny
Phylogenetics has been used in one form or another for over a century and a half. Darwin drew a phylogenetic tree in his notebook in 1837, with the caveat “I think” written in the margin.2 The first use of the word phylogenetics appeared in 1921 in a paper on New Zealand stone flies.3 Phylogenetics was not completely codified into a classification system until 1950, but the idea of a branching tree of life was widespread long before then.
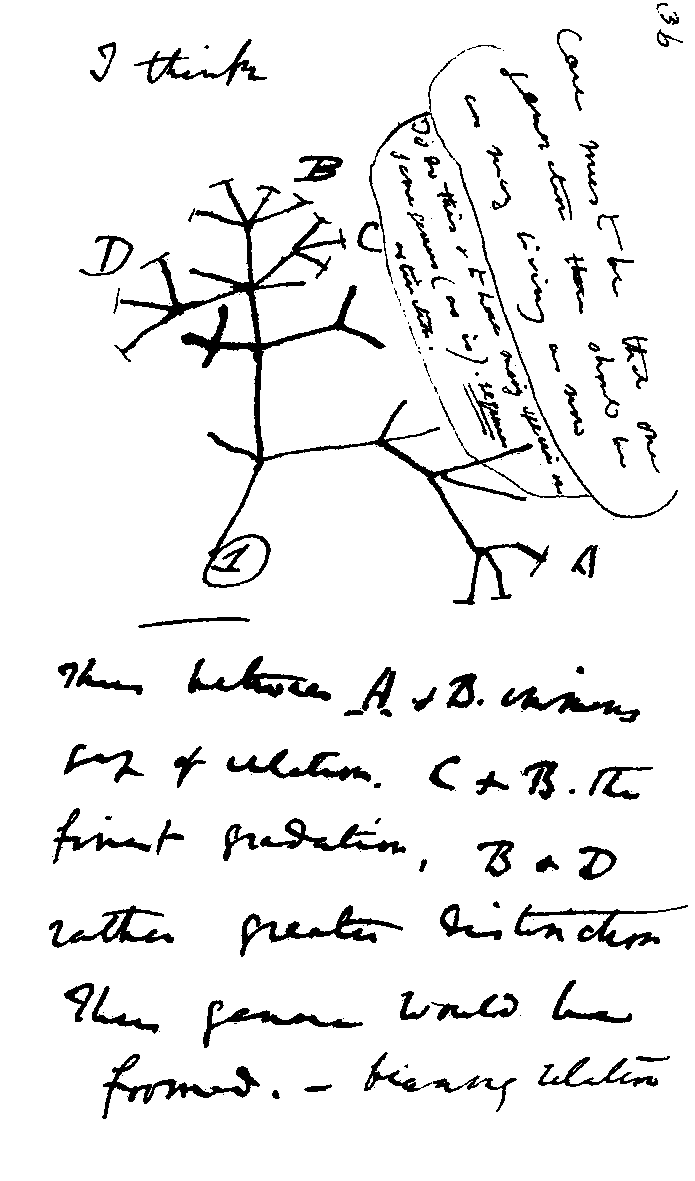
Darwin's tree drawn in his notebook with the important caveat "I think" in the upper left. Charles Darwin, Public domain, via Wikimedia Commons.
Darwin drew a phylogenetic tree in his notebook in 1837, with the caveat “I think” written in the margin.
The man who would finally codify phylogenetics into a form of biological systematics was Willi Hennig. Hennig was a German entomologist who fought for Germany during World War II and was wounded in Russia. Prior to the war in 1933, he had voluntarily joined the SA, Hitler’s brownshirts notorious for brutalizing and murdering political opponents and later Jews.4 Hennig’s level of involvement in the domestic terrorist organization is unknown, but his involvement was voluntary and indicates at least some sympathy for Nazi ideas. It is possible that Hennig’s views on Nazism changed over time, however, as his personal papers do not contain much allusion to Nazism, nor did he close personal letters with the expected salute to Hitler. Despite his ties to the brownshirts, Hennig was ignored after the war, likely due to his focus on insect studies rather than anything that could be construed as martial. Based on his studies of insects and his Darwinist worldview, Hennig attempted to solve the taxonomic riddles of his day by proposing phylogenetics.
Hennig’s book was translated into English in 1966. He made it abundantly clear that his classification was based on his belief in Darwinism. While describing the starting point of the systematist, Hennig wrote, “A much better point of departure is to recognize that evolution is a fact and that its course and the conformities to law that control it must be investigated.”5 In other words, Hennig assumed that evolution must be true and built his system of classification on that assumption.
A Historical Science
Phylogenetics as a whole is more about history than it is about the present. The goal is to reconstruct the evolutionary history of the past. “The method proposed to recover ‘historical structure’ is that of phylogenetics systematics.”6 Evolutionists themselves long have believed that evolutionary biology is historical rather than empirical. One author wrote in 1983, “In essence, an evolutionary explanation is a historical interpretation, and so may not yield predictions which can be tested now.”7 More recently, Jonathan Losos, a textbook author and herpetologist, echoed that sentiment. “But evolutionary biology is a historical science. Like astronomers and geologists, we evolutionary biologists try to figure out what happened in the past. And like historians, we are bedeviled by the asymmetry of time’s arrow—we can’t go back in time to see what happened. Moreover, evolution occurs notoriously slowly, seemingly making it impossible to watch as it occurs.”8 In other words, evolutionary explanations, such as phylogenetics, are historical by default.
Some evolutionists have gone even further and claimed that the only nonhistorical part of the taxonomic hierarchy is the presence of existing species. “Whereas supraspecific taxa are collections of lineages, species are the lineages themselves. These lineages are placed in higher groupings based on hypothesized past linkages. Thus, natural higher taxa are purely historical constructs whose sole existence depends on how accurately they document the historical unfolding of lineage splitting (speciation).”9 This means that the entire classification system as built by phylogenies are historical constructs that rely on the accuracy of phylogenies to be true. If the underlying assumptions of phylogenies are inaccurate—and some inaccuracies will be demonstrated below for certain evolutionary phylogenies—then the historical structure falls apart. It is possible to build a phylogeny which matches with Scripture. However, this possibility does not necessarily mean that all phylogenies are accurate. Everything depends on the assumptions used to build and interpret the phylogeny.
Phylogenetic Circularity?
To this day, mainstream phylogenetics continues to assume evolution to be true to form their classifications. However, the exact method they choose to use is still highly controversial. Some evolutionists have recognized the inherent circularity of assuming evolution to prove evolution: “[M]y argument is that the theory of macroevolution is corroborated by evidence from systematics (i.e., comparative anatomy, paleontology, biogeography, and more, recently, comparative biochemistry). Therefore, the a priori assumption of descent with modification fails to provide independent ontological support for systematics. If ‘the background knowledge of descent with modification’ underlying cladistics is not testable by independent means, it would seem to be more of a metaphysical First Principle like vitalism or orthogenesis than a component of a Popperian hypo-thetico-deductive approach.”10 This author, an evolutionist himself, believes that many evolutionists are appealing to the metaphysical to support their story, rather than rigorous empirical science. Other evolutionists, of course, reject this notion, believing they are dealing with empirical data.
To this day, mainstream phylogenetics continues to assume evolution to be true to form their classifications. However, the exact method they choose to use is still highly controversial.
Other evolutionists believe that the circularity is not an issue. Noted biologist E.O. Wiley wrote, “The formalism of taxonomy must give way to the perceived realities of evolution.”11 In other words, evolution must overrule anything systematics produces that does not agree with the dogma. Another pair of authors wrote, “Hence the approach advocated by Hennig is not circular because homologies, which indicate phylogenetic relationships are determined without a priori reference to a phylogeny, while homoplasies, which are inconsistent with phylogeny, are determined by reference to a phylogeny”12 (emphasis theirs). Their argument is that homologous characters indicate phylogenetic relationships that are not determined by phylogenies. However, determining characters which were absent in the common ancestor but present in the studied species, the homoplasies are discovered by a phylogenetic tree. The idea is that phylogenetics is not circular because it does not attempt to determine homologous traits by appealing to homoplaisic traits and vice versa. Of course, it is determining homoplasies in a post-hoc fashion, which these researchers even admit only a few pages earlier in their book.13
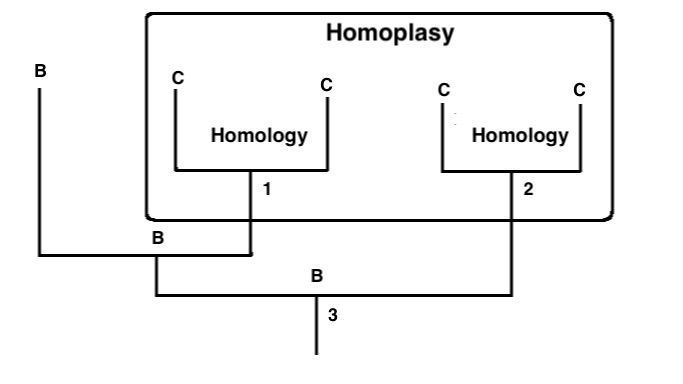
Homology and Homoplasy demonstrated on a phylogenetic tree. Similar traits that share ancestry are homologous. Similar traits that do not share ancestry are homoplaseous.
Unfortunately for the evolutionary mindset, the homologous/homoplaisic argument against circularity does not address the issue of phylogenies assuming evolution and then proving evolution. In fact, one evolutionist points out that phylogenetics requires evolution as its philosophical foundation. “Cladism needs evolution in its techniques, although not actually to operate them in the day-to-day practical sense. It needs evolution in its philosophy. The theory of evolution is necessary–not just desirable or decorative–in the justification of cladism.”14 In other words, according to evolutionists themselves, phylogenetics requires evolution to be true in order to even function. This is posturing, to an extent, because it is possible to construct phylogenies that do not require molecules-to-man evolution and simply show how different organisms are, rather than assuming the difference between them is due to ancestry. However, it is ironic given the statement from another phylogeneticist that “although a central tenet of modern biology, evolutionary theory is not a ‘fact,’ but a causal explanation for observed regularity of character distribution.”15
Unfortunately for the evolutionary mindset, the homologous/homoplaisic argument against circularity does not address the issue of phylogenies assuming evolution and then proving evolution.
Character Selection
In order to construct a phylogenetic tree, evolutionists compare characters between organisms. Philosophically, they are making the assumption that similarity is equivalent to ancestry and are interpreting their results accordingly. “We would stress that an evolutionary model is NOT just the mechanism of character change; it includes the assumption of searching for a tree as the underlying relationship among the taxa.”16 These researchers believe that character change comes from evolution, and hence looking for them should yield a phylogenetic tree.
Evolutionists believe characters are a major part of the evolutionary process. “Characters can also be defined theoretically. They can be divided into evolutionarily stable characters which (we infer) do not change during evolution, and evolutionarily labile ones, which change more often. Or they can be divided into evolutionarily ‘ancestral’ characters and evolutionarily derived ones, which are earlier and later evolutionary states of the same character; fins and limbs are an example.”17 Evolution is believed to produce new characters as part of the process. These characters are automatically more advanced than the “primitive” characters that proceeded them. They are more advanced because they are the result of evolution, not because they actually are more advantageous to the organism.
What then are these characters that evolutionists use to build their trees? Wiley defined characters: “A character is a feature of an organism which is the product of an ontogenetic or cytogenetic sequence of previously existing features, or a feature of a previously existing parental organism(s). Such features arise in evolution by modification of a previously existing ontogenetic or cytogenetic or molecular sequence.”18 Thus, according to Wiley, a character can be an external or phenotypic trait generated either as an adult or during development or a genetic sequence.
Character choice is very important when building a phylogenetic tree. In the evolutionists’ minds, not all characters are created equal. Some have more value than others, depending on the study. Wiley makes this point: “The kind of quantitative characters an investigator will select depend on the purpose of the study.”19 For example, if they are trying to determine how two birds are related to one another, feathers would be a bad character to use. One author illustrated their mindset very well. “So taxonomists are faced with a choice. They have to choose some characters rather than others. They might, of course, pick characters arbitrarily and define arbitrary groups, but in practice they do not. They generally try to choose particular kinds of characters; and their choice is dictated by taxonomic principle. . . . Taxonomists have been forced to develop a philosophy, in addition to the practical task of defining groups by the fact that different characters disagree.”20 What taxonomic principle do they mean? “All phylogenetic cladists adopt the basic premise that life has evolved.”21 Character selection depends on the purpose of the phylogeny, and often the overarching purpose is the resolution of purported evolutionary relationships. For example, the red panda was originally classified as a raccoon, then reclassified as a bear because it had a pseudo-thumb like a panda. It has since been reclassified again into its own group based on genetic data. Each of these changes was based on selection of characters. Thus, it is incredibly important to select the correct characters to correctly classify anything.
Character choice is very important when building a phylogenetic tree. In the evolutionists’ minds, not all characters are created equal. Some have more value than others, depending on the study.
A Perfect Tree?
Evolutionists are open about the fact that they cannot show any phylogenetic tree to be fully accurate. Within their own community, they repeatedly make this point: “Since phylogenetic trees are hypotheses and not ‘facts,’ they are dependent upon both the quality and quantity of data that support them.”22 This author recognizes that phylogenetic trees are dependent heavily on how good their data is. Another author concurs, noting, “Evolutionary classifications are always subject to improvement.”23 A third goes even further, making a shocking claim about the nature of phylogenetics: “Probably no phylogenetic cladogram, no matter how it is constructed, can be totally ‘proved’ or ‘falsified.’”24 This has led one scientist to propose a complete abandonment of falsification as a criterion for phylogeny.25 Obviously, comparing phylogenies against historical records can help, but there is a great deal of data which has been lost over time, making it very difficult to falsify a phylogenetic tree.
The problem becomes more difficult given a remarkable statement published in the journal Evolutionary Biology in 2016: “Cladograms lack the causal details of the different hypotheses implied by those diagrams to make testing feasible, and falsification has been shown to be problematic for historical sciences.”26 If something cannot be falsified, as authorities on the topic admit, then it is not part of empirical science. And according to this author, not only is phylogenetics not observable, it is not testable either! Being testable is absolutely necessary for something to be empirical science. This may be why one scientist, a phylogenetic researcher himself, wrote, “I do not think that claims made by cladists themselves can always be taken at face value.”27 Again, the important takeaway is not necessarily that the underlying equations are false, but that the interpretations of the results are not falsifiable in an evolutionary millions-of-years paradigm.
Even worse, the same models of phylogenetic trees which evolutionists use for animals do not work for plants. Yet Verne Grant, perhaps the most well-regarded botanist of the twentieth century, wrote, “The phylogeny of many or most groups of higher plants which have been studied carefully in the field and laboratory is seen to be reticulate rather than exclusively dichotomous and branching. The model of the phylogenetic tree cannot be carried over from zoology into botany without misrepresenting the relationships in many instances. In all major groups of higher plants, we have to deal with phylogenetic networks rather than phylogenetic trees.”28 Grant recognized the impossibility of importing zoology into botany when it came to systematics. In fact, if you read him critically, he almost seems to be describing a creation orchard rather than an evolutionary tree. Obviously, plants have more options for genetic change available, which complicates building a tree.
To illustrate how subjective this is, examine phylogeny output 1–3 below. We have used a phylogeny builder online using DNA sequences from mustelids and other animals, both supposedly related and unrelated according to evolution. These tools are also used by evolutionary biologists to perform their phylogenies.29,30,31,32,33,34,35 The data is freely available online from NCBI. We used the same type of data as evolutionists themselves have used to classify Mustelidae.36 We are aware that using a single gene such as we have done can yield some strange results, but the evolutionists themselves have published it in peer-reviewed journals, so it seems appropriate to critique it here. Building lineages works much better with full mtDNA sequences.37

Phylogeny output figure 1. A basic phylogeny of Mustelidae using full mtDNA sequences. Note that the beech marten does not group with the rest of its genus.

Phylogeny output figure 2. First phylogeny with two added taxa.
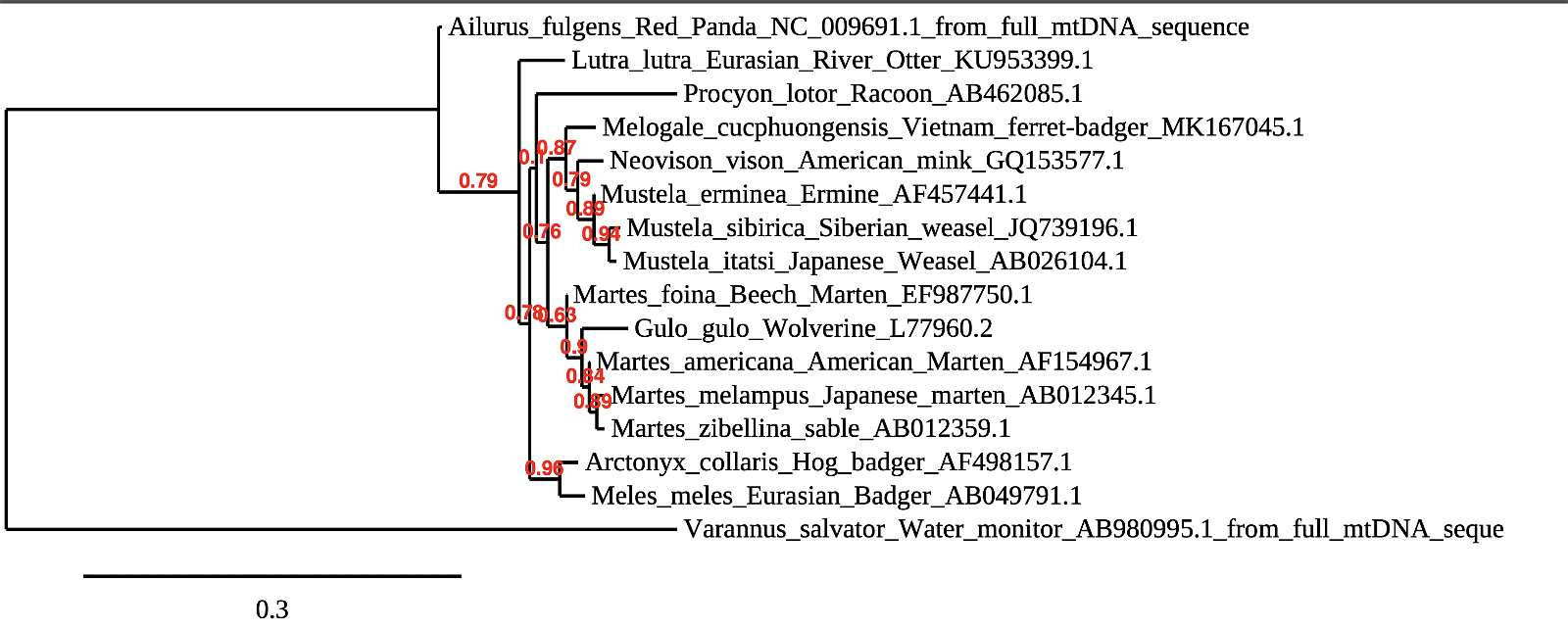
Phylogeny output figure 3. First phylogeny with three added taxa. Note how the tree changes with added taxa.
Phylogeny samples 1–3 illustrate how subjective the results of such phylogenies can be. We used full sequences of the mtDNA gene cytochrome b from Mustelids. A few absurdities pop up immediately, particularly the beech marten being on the same branch as the otters and wolverines, but not the other three members of its genus in this analysis. The second phylogeny adds raccoons and red pandas to the tree, which are supposed to be the Mustelids’ closest relatives. Red pandas mysteriously are considered more closely related to otters than otters are to other Mustelids. As seems common, the beech marten is recovered as being more closely related to wolverines than to other martens. The third phylogeny figure introduces a monitor lizard, the water monitor, a close relative of the Komodo dragon. This figure does recover the monitor lizard in its own group, but other anomalies exist. The beech marten continues to nest away from the rest of its genus, while the raccoon is recovered as more closely related to weasels than the otter, which is in the same family as weasels.
Repairing a Broken System
Mainstream phylogenetics fails because it is built on a false foundation. The secular phylogenies require evolution. “Evolution results in natural groups because all evolutionary change takes place in the same phylogenetic tree: all changes in all characters must have taken place within the same pattern of lineages.”38 Evolution and phylogenetics are intertwined like two strands of a vine. Since evolution is false, so must also be mainstream phylogenetics. Like any other false system, this one can be reduced to absurdity by applying its own logic to itself. It is purely circular to assume evolution to demonstrate evolution. Yet phylogenetics is still passed off as evidence for their dogma.
The problem is that nested hierarchies are in a zone of overlap between creation and evolution. Both creationists and evolutionists predict that there will be a certain level of continuity of life. To evolutionists, the homologies they observe are evidence of common ancestry. Thus, when they interpret phylogenies, they assume that similar organisms are more closely related. This, however, is not always the case, hence why evolutionists frequently appeal to convergent evolution. This can be illustrated by models of vehicles. When humans design things, they design them in nested hierarchies. SUVs made by different manufacturers will share more in common than a truck and an SUV made by the same manufacturer because they share a common purpose, not because they share a common lineage. This is also true of the biological world. Similar structures are not necessarily homologous in the evolutionary sense. They may simply share similar functions. There are only so many ways to design, say, an arm in an optimal fashion for function. Therefore, there will always be homology between organisms that perform similar functions, related or otherwise. And wherever there is homology, there will be an evolutionist claiming the organisms are related, whether they are or not.
Mainstream phylogenetics fails because it is built on a false foundation. The secular phylogenies require evolution.
“We all have our biases. The wonder is that any of us do come close to the truth about the past instead of just constructing a plausible story,”39 said Mary Pickard Winsor an evolutionist writing in the context of phylogenetics. She is, of course, completely correct. Everyone has a bias, and that bias influences how we view the past. What she misses is that there is an unbiased eyewitness account of the past: God’s Word. The Bible presents the only eyewitness account of the origin of variety on earth, and it does not talk about a universal common ancestor, the phylogenetic tree of evolution. Instead it talks about an orchard, kinds of organisms, mostly at the family level of classification, which God created on days three, five, and six. Until systematists are willing to take God at his Word, they will be unable to provide a coherent classification system.
Answers in Depth
2020 Volume 15
Answers in Depth explores the biblical worldview in addressing modern scientific research, history, current events, popular media, theology, and much more.
Browse VolumeFootnotes
- Neil A. Campbell and Jane B. Reece, Biology (San Francisco: Pearson, 2005), 491.
- Charles Darwin, “Notebook B: Transmutation of species (1827–1838)” transcribed by Kees Rookmaaker (accessed November 22, 2019), http://darwin-online.org.uk/content/frameset?pageseq=1&itemID=CUL-DAR121.-&viewtype=text.
- R.J. Tillyard, “A New Classification of the Order Perlaria” The Canadian Entomologist 53, no. 2 (1921): 35–43, https://www.cambridge.org/core/journals/canadian-entomologist/article/new-classification-of-the-order-perlaria/19A45868AC05A1048D88466C6210608D.
- Michael Schmitt, From Taxonomy to Phylogenetics: Life and Work of Willi Hennig (Boston: Brill, 2013), 30.
- Willi Hennig, Phylogenetic Systematics, trs. D. Dwight Davis and Rainer Zangerl (Chicago: University of Illinois Press, 1979), 234.
- Olivier Rieppel, “Species and History” in Models in Phylogeny Reconstruction, eds. Robert W. Scotland, Darrell J. Siebert, and David M. Williams (Oxford: Clarendon Press, 1994), 33.
- Oliver Mayo, Natural Selection and Its Constraints (London: Academic Press Inc. Ltd, 1983), 1.
- Jonathan Losos, Improbable Destinies: Fate, Chance, and Future of Evolution (New York: Riverhead Books, 2017), 20..
- E. O. Wiley, Phylogenetics (New York: John Wiley & Sons, 1981), 26.
- Andrew V. Z. Brower, “Evolution Is Not a Necessary Assumption of Cladistics,” Cladistics 16, no. 1 (2000): 143–154, https://onlinelibrary.wiley.com/doi/pdf/10.1111/j.1096-0031.2000.tb00351.x.
- Wiley, 26.
- Daniel R. Brooks and Deborah H. McLennan, Phylogeny, Ecology and Behavior (Chicago: University of Chicago Press, 1991), 25–26.
- Ibid, 14.
- Mark Ridley, Evolution and Classification (London: Longman Group Limited, 1986), 162.
- Rieppel, 1994, 38.
- David Penny, Peter J. Lockhart, Michael A. Steel, and Michael D. Hendy, “The Role of Models in Reconstructing Evolutionary Trees” in Models in Phylogeny Reconstruction, eds. Robert W. Scotland, Darrell J. Siebert, and David M. Williams (Oxford: Clarendon Press, 1994), 33.
- Ridley, 2–3.
- Wiley, 116.
- Ibid, 340.
- Ridley, 2.
- N.R. Scott-Ram, Transformed Cladistics, Taxonomy and Evolution (Cambridge: Cambridge University Press, 1990), 179.
- Brooks and McLennan, 31.
- Ridley, 27.
- W.H. Wagner Jr., “Origin and Philosophy of the Groundplan-divergence Method of Cladistics” in Cladistic Theory and Methodology, ed. Thomas Duncan and Tod F. Stuessy (New York: Van Nostrand Reinhold Company, 1985), 166.
- Lars Vogt, “Popper and phylogenetics, a misguided rendezvous.” Australian Systematic Botany 27, no.2 (2014) 85–94, http://www.publish.csiro.au/SB/pdf/SB14025.
- Kirk Fitzhugh, “Phylogenetic Hypotheses: Neither Testable Nor Falsifiable” Evolutionary Biology 43, no. 2 (2016) 257-266, https://link.springer.com/article/10.1007/s11692-016-9381-8.
- David L. Hull “Cladistics Theory: Hypotheses That Blur and Grow” in Cladistics ed. Thomas Duncan and Tod F. Stuessy (New York: Columbia Univeristy Press, 1984), 16.
- Verne Grant, Plant Speciation (New York: Columbia University Press, 1981) 481.
- Guillaume Blanc et al., “BLAST-EXPLORER Helps You Building Datasets for Phylogenetic Analysis” BMC Evolutionary Biology 10, (2010): https://bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-10-8.
- Olivier Gascuel et al., “Phylogeny.fr: Robust Phylogenetic Analysis for the Non-specialist” Nucleic Acids Research 36, suppl. 2 (2008): W465–W469, https://academic.oup.com/nar/article/36/suppl_2/W465/2505761.
- Robert C. Edgar, “MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput” Nucleic Acids Research 32, no. 5 (2004): 1792–1797, https://academic.oup.com/nar/article/32/5/1792/2380623.
- J. Castresana, “Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis” Molecular Biology and Evolution 17, no. 4 (2000): 540–552, https://academic.oup.com/mbe/article/17/4/540/1127654.
- Stephane Guindon and Olivier Gascuel, “A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood.” Systematic Biology 52, no. 5 (2003): 696–704, https://academic.oup.com/sysbio/article/52/5/696/1681984.
- Maria Anisimova and Olivier Gauscel, “Approximate Likelihood-Ration Test for Branches: A Fast, Accurate and Powerful Alternative” Systematic Biology 55, no. 4 (2006): 539–552, https://academic.oup.com/sysbio/article/55/4/539/1675125.
- Francois Chevenet et al., “TreeDyn: Towards Dynamic Graphics and Annotations for Analyses of Trees” BMC Bioinformatics 7 (2006): https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-7-439.
- Josep Marmi et al., “Phylogeny, Evolutionary History and Taxonomy of the Mustelidae Based on Sequences of the Cytochrome b Gene and a Complex Repetitive Flanking Region” Zoologica Scripta 33, no. 6 (2004): 481–499, https://onlinelibrary.wiley.com/doi/abs/10.1111/j.0300-3256.2004.00165.x.
- Nathaniel Jeanson, “Mitochondrial DNA Clocks Imply Linear Speciation Rates Within ‘Kinds,’” Answers Research Journal 8 (2015): 273–304, https://assets.answersresearchjournal.org/doc/v8/mitochondrial-dna-clocks-linear-speciation-rates.pdf.
- Ibid, 19.
- Mary Pickard Winsor, “The Lessons of History” in Models in Phylogeny Reconstruction, eds. Robert W. Scotland, Darrell J. Siebert, and David M. Williams (Oxford: Clarendon Press, 1994), 2.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis