Reproductive Thievery in Salamanders?
Abstract
Two evolutionary hypotheses have been put forth to explain the Jefferson complex and the various modes of reproduction that may have taken place.
Introduction
The first rainy nights of early spring produce an amazing phenomenon that inspires awe in my students every year. It is the first time they witness the great salamander migrations of south central New York. Many of these amphibians are mole salamanders in the Ambystomatidae family, which are characterized by stout bodies and round heads.1 They spend most of the time underground, and, after having survived a harsh winter, they emerge by the thousands and make a beeline for breeding pools. They tend to be locally rare and, depending on their geographical location, may be classified as a species of concern, threatened, or endangered.2
Though details of mating behavior differ between species, they tend to choose breeding pools that are free of fish, and males perform complex mating rituals to attract females. A male grasps her, usually behind the forearms, and undulates his tale while producing chemical signals from the cloaca that may be important in sex recognition and distinguishing each other as a species. Afterward, he deposits a spermatophore on the bottom of the pool. Since males do not have the genitalia needed to deposit sperm into a female, they produce a spermatophore which consists of a gelatinous “bag” topped with a sperm cap containing seminal fluid. If the female is interested, she will use her cloaca to pinch off the sperm cap and incorporate the sperm so that internal fertilization can take place. She deposits a jelly-like egg mass onto underwater vegetation, and both will leave the pool to return to their subterranean burrows until next spring.
The Jefferson Complex of Salamanders
Complicating their reproductive life histories is the existence of a large number of females called unisexuals. Male unisexuals exist but they are rare and probably infertile.3 Unisexuals are thought to be products of past hybridization with other salamanders in the family4 and can be found in breeding pools from the Great Lakes region to the deciduous forests of Eastern North America. Individuals have uneven mixtures of genes from multiple salamander species and, until recently, four species were thought to be involved: the Jefferson (J) (Ambystoma jeffersonianum), blue spotted (L) (A. laterale), small mouth (T) (A. texanum), and tiger salamanders (Ti) (A. tigrinum). A fifth species, the stream-side salamander (A. barbouri), has been included with the others and are collectively called the Jefferson complex of salamanders.
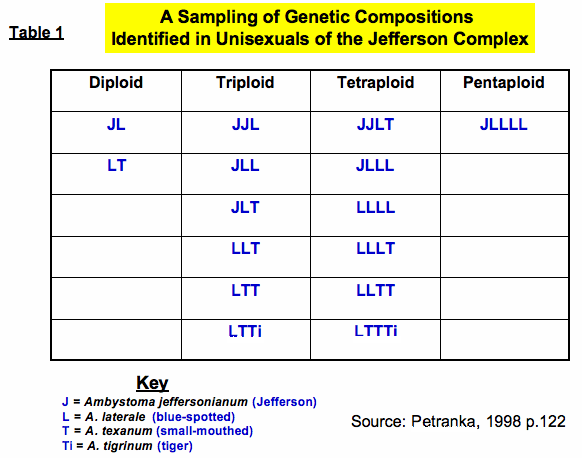
Normally adult salamanders have diploid (2n = 28) body cells where one set of chromosomes (n = 14) comes from mom and the other (n = 14) comes from dad. Interestingly, the unisexuals of the Jefferson complex can be diploid hybrids or polyploid. Polyploidy is a rare and often lethal condition in animals. It occurs when more then two sets of chromosomes become incorporated into cells. Researchers have identified triploid (3n), tetraploid (4n), and pentaploid (5n) salamanders in this complex. Triploids are fairly common in any given population while tetraploids and pentaploids are found in much lower frequency.1 At first these creatures were formally named, but because the genetic story is far more complex, they are currently identified based on their genetic composition (see Table 1). Triploids have intermediate morphologies, are mostly female, and have larger eggs, cells, and cell nuclei, but deposit smaller clutch sizes than the purebred species.1 They are difficult to recognize in the field and are best identified genetically by analyzing chromosome numbers, microsatellite DNA, mitochondrial DNA (mtDNA), and allozymes (variant forms of the same enzyme). Recent research has produced a non-invasive means to rapidly distinguish between unisexual and bisexual individuals using a small tissue sample and analyzing the cytochrome b region of mtDNA.5
Over the years, researchers have hypothesized that unisexuals were products of various forms of reproduction that include parthenogenesis, hybridogenesis, gynogenesis, and unique forms of sexual reproduction.6 Parthenogenesis is a form of animal asexual reproduction where eggs develop without sperm and offspring are produced from unfertilized eggs.
Hybridogenesis is an unusually rare type of reproduction, is not completely asexual, and may have been identified in some frogs.7 For example, if hybridogenesis was happening, a Jefferson salamander (J) may hybridize with a blue-spotted salamander (L) and produce hybrid offspring (JL). As adults, the hybrids could backcross with purebred Jefferson males. During the larval stage of the new offspring, the Jefferson male genome is not incorporated, leaving the original (JL) female genetics intact for the next generation. In effect, the donor father’s sperm is taken but discarded early in larval development.
Gynogenesis occurs when a sperm from a diploid male stimulates a triploid female egg to develop, but never gets incorporated into the new zygote. Triploids (3n), and on rare occasions tetraploids (4n) and pentaploids (5n), are produced through ploidy elevation.
Sexual reproduction can produce elevated ploidy levels. Sperm may contribute their chromosomes into unreduced 2n eggs, 3n eggs or 4n eggs and produce offspring of a higher ploidy level. Therefore, diploid females (2n) may produce triploid offspring, triploid females (3n) may produce tetraploid (4n) offspring, and tetraploid females (4n) may produce pentaploid offspring (5n).
Origins of the Jefferson Complex
Two evolutionary hypotheses have been put forth to explain the Jefferson complex and the various modes of reproduction that may have taken place.1 One hypothesis is that millions of years ago there was a small number of hybridization events followed by diploid hybrids backcrossing with purebreds and producing unisexual polyploids (JJL, JLL). Gynogenesis then ensues from generation to generation.
The second possibility of unisexual origins is the hypothesis that they have arisen on multiple occasions rather than one or a small number of times. Continued gene exchange between unisexuals and diploids continued at low rates. Evidence given for this hypothesis was that diploid (JL) and triploid (JJL and JLL) salamanders can arise from the same egg masses.
Because of the surprising genetic uniformity in unisexual species, except for A. texanum, many researchers tended to favor the first hypothesis because the above suggests that the complex was produced by a small number of events rather than at high rates on multiple occasions.8
The Mystery Deepens
Current data suggest that unisexual Ambystoma are not parthenogenetic because if they were, repeating nucleotide sequences of a few base pairs, called microsatellite DNA, should be the same among individual salamanders and they are not. Microsatellite DNA is used to determine kinship between individuals. In one study only 2/26 of the unisexual egg masses and 2/30 of unisexual adults had the same genetic sequences.6
There is also no evidence that hybridogenesis is taking place because it has not been demonstrated that a common genome is being transmitted from one generation to the next. Furthermore, what seems to tie the complex together genetically is a fifth species known as the stream-side salamander (A. barbouri). MtDNA sequence data is quite uniform in unisexuals and suggests they descended from a common maternal ancestor of A. barbouri that diverged 3-4 mya, according to “molecular clock” calculations.6 Historically this salamander was considered conspecific with the small-mouth salamander (A. texanum - T). Their populations don’t tend to overlap, but sometimes they do, and hybridization has been documented. The stream-side differs from the small-mouth in breeding habitat, mode of egg deposition, egg size, clutch size, larval morphology, larval behavior, and teeth, and for these reasons are considered separate species.9
What, then, is going on? Recent analysis of allozymes and microsatellite DNA suggest that unisexual females are “stealing” male sperm from syntopic species (differing species in the same place at the same time) and incorporating the male genomes into their diploid/polyploid nuclei. In other words, these salamanders may be participating in a strange form of reproduction known as kleptogenesis, which may be unique in the animal world. Apparently these female kleptogens have a flexible enough chemistry to incorporate several different species of male sperm. Gynogenesis and/or ploidy elevation may further the process to produce nuclear genomic hybrids. This explanation seems to explain the data well, including unisexual uniformity of unrelated mtDNA sequences matching A. barbouri.
The Jefferson Complex with a Creation Perspective
The Jefferson Complex of salamanders is a great example of the difficulty with defining a species precisely. Though the general concept of species as a recognizable organism is useful in biosystematics and fieldwork, historically it has been arbitrarily defined, and a consensus definition is wanting. One definition often cited is a taxonomic group that can interbreed, but there are many others. The Jefferson complex of salamanders complicates how a species might be defined.
Baraminology is a creationist method of biosystematics where the goal is to define real groups of organisms based on the created “kinds” of Genesis 1.10 Creationist researchers have begun to analyze the Ambystomatidae family. Using statistical analysis and documenting the ability of hybridization in bisexuals, preliminary results have classified them as a monobaramin.11 A monobaramin is defined as the group of known organisms that share biologically meaningful similarity with one other.12 Except for the time element, and from a young earth creation perspective, past hybridization events and kleptogenesis are not inconsistent hypotheses for the origins of Jefferson complex unisexuals.
However, if we take the underlying differences between an evolutionist and creationist to a deeper level, it is not the science that is being argued, but the worldviews of the researchers. If we can differentiate the evolutionary worldview from the biblical worldview we can examine assumptions at a deeper level.
The evolutionary interpretation tells of an ancestral female stream-side salamander diverging 3-4 million years ago. This interpretation often assumes several things. First, it assumes that deep time has happened and that millions and billions of years have passed since the random origin of first life. Second, it assumes the idea of neutral evolution where it is believed that the majority of DNA is junk and without function.13 As more data are revealed, the concept of non-functional DNA is becoming harder to demonstrate. Third, this assumes that a “molecular clock” exists. The 3-4 million year divergence is based on the molecular clock hypothesis which assumes that we can determine the mutation rate in mtDNA. This hypothesis, in turn, assumes that neutral evolution is true and the majority of DNA is without function. However, these concepts make numerous other assumptions which have all been contradicted in the evolutionary literature.14, 15 Even many evolutionists are questioning the notion that a molecular clock exists.
In contrast, biblical creationists recognize the Bible as the Word of the Creator and have a different set of assumptions. The Bible clearly reveals that life was deliberately created and designed by God thousands of years ago. Destruction was brought upon the planet by a great Flood 4500 years ago. Land vertebrate air breathers observed today are assumed to have diversified from the originals brought aboard the ark. Mole salamanders, being terrestrial in the adult form, could have been easily cared for. As Christians who believe God and His Word, we assume God has created His creatures to persist. In a fallen world these creatures are predicted to have the genetic machinery to respond to changing environments and have all sorts of ingenious methods for accomplishing what the Creator has given them to do. This understanding suggests that the Ambystomatidae family probably started from at least a single pair, since the flood, no more than 4500 years ago.
There is still much to learn about the Jefferson salamander complex. Just how they diversified and the non-random, programmed mechanisms capable of responding to changing environments are still mysteries, but they continue to inspire exciting venues for creationist research. These salamanders are an amazing testament to the power and creativity of our Creator and Savior Jesus Christ. Though my students may not recognize the true source of their awe as they beheld these creatures and helped them cross the road, my prayer is that one day they will meet and trust the One who was the basis of their awe, the Author of Life and Savior of their souls.
Footnotes
- Petranka, J.W. 1998. Salamanders of the United States and Canada. (Washington D.C.: Smithsonian Institution Press), pp. 122–129.
- Julian, S.E., King, T.L. and Savage, W.K. Novel jefferson salamander, Ambystoma jeffersonianum, microsatellite dna markers detect population structure and hybrid complexes. Molecular Ecology Notes 3:95–97.
- Bogart, J.P., Klemens, M.W. 2008. Additional distributional records of Ambystoma laterale, A. jeffersonianum (amphibia: caudate) and their unisexual kleptogens in northeastern North America. American Museum Novitates 3627:1–58.
- Gibbs, J., Breisch, A.R., Ducey, P.K., Johnson, G., Behler, J.L., Bothner, R.C. 2007. Amphibians and Reptiles of New York State: Identification, History, and Conservation. (New York: Oxford University Press) pp. 62–63.
- Noël, S., Dumoulin, J., Ouellet, T., Galois, P., and Lapointe, F-J. 2008. Rapid identification of salamanders from the Jefferson complex with taxon-specific primers. Copeia 1:158–161.
- Bogart, J.P., Bi, K., Fu, J., Noble, D.W.A., and Niedzwiecki, J. 2007. Unisexual salamanders (genus Ambystoma) present a new reproductive mode for eukaryotes. Genome 50:119–136.
- Beerli, P. 1995. Hybridogenesis in water frogs. http://www.tolweb.org/notes/?note_id=579 accessed 2 May 2010.
- Petranka, p. 123
- Petranka, pp. 40–46.
- Wood, T. and Murray, M.J. 2003. Understanding the Pattern of Life (Nashville, TN: Broadman & Holman Pub.), p. 3.
- Brophy T., and Kramer, P.A. 2007. Preliminary results from baraminological analysis of the mole salamanders (Caudata: Ambystomatidae). Occasional Papers of the BSG 10:10–11.
- Wood, T.C., Wise, K,. Sanders, R., and Doran, N. 2003. A Refined Baramin Concept. Occasional Papers of the BSG 3:1–14.
- Carter, R. 2009. The slow, painful death of junk dna. http://creation.com/junk-dna-slow-death accessed 6 May 2010.
- Carter, R. 2009. The neanderthal mitochondrial genome does not support evolution. Journal of Creation 23(1):40–43. http://creation.com/images/pdfs/tj/j23_1/j23_1_40-43.pdf accessed 6 May 2010.
- Carter, R. 2007. Mitochondrial diversity within modern human populations. Nucleic Acids Research 35(9):3039–3045. http://nar.oxfordjournals.org/cgi/reprint/35/9/3039 accessed 6 May 2010.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis


