The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
Charles Darwin is well known as having popularized the concept of natural selection. Despite widespread teaching of the topic in biology related contexts, it is actually controversial among scientists. This is true in both evolutionary and creationary circles.
To assess the role natural selection plays in our world, it must first be defined in a scientifically rigorous manner. Mathematical tools can help in assessing its role; comparison with principles known from artificial selection can be informative as well. With various methods of mathematical modeling and statistical tests, there are important assumptions that should be recognized to avoid premature conclusions about the results. Finally, field observation provides important data that needs to be considered.
Examination of each of these areas leads to several conclusions. Natural selection has not played a significant positive role in the history of life on earth for sexually reproducing animals such as mammals and birds. Statistical tests, which are a valid means to screen for natural selection and comprise the one line of evidence that often seems to support its occurrence in adaptation, have not been validated; many of the non-random patterns detected probably have little to do with the action of natural selection. Naturalistic assumptions, which include assuming meiotic drive (which distorts Mendelian inheritance) is a random mechanism, appear to have hindered for decades our understanding of how creatures adapt. A reasonable creationary prediction is that mechanisms such as biased gene conversion (one form of meiotic drive) will turn out to be designed to alter the frequency of alleles in a way that aids in adaptation of the population.
Keywords: evolution, natural selection, adaptation, speciation, mutations, population genetics, statistical tests, genetic drift, finches
Introduction
Natural selection is a concept popularized by Charles Darwin as a naturalistic explanation for the variety we see in life today and why so many creatures seem perfectly adapted to their environment. He recognized that there is variation in living things. He argued that some traits may be better suited for a particular environment than others. Those individuals with more advantageous traits would be more likely to survive and reproduce. In this way those desirable traits would be passed on to their offspring and become more common in the population. Given enough time, he believed this could explain how one kind of animal (for example, a dog) could have descended from a very different kind of animal (for example, an invertebrate).
Given the importance of natural selection within the popular-level neo-Darwinian explanations of evolution, it is a surprise to many to find out that the concept is controversial among evolutionists.1 There are atheistic scientists who propose that Darwin was wrong about natural selection and that the concept isn’t even scientific (Fodor and Piattelli-Palmarini 2010). Others, while recognizing significant problems in how natural selection is often portrayed, feel that it still has an important role in accounting for how creatures become adapted to their habitats (Endler 1986).
Natural selection hasn’t just been considered important in the evolutionary model. Creationists recognize that there is incredible variety seen in species derived from the same created kind. While pointing out the insufficiency of natural selection to account for the evolutionary changes necessary for molecules-to-man evolution, it is common for creationists to affirm natural selection as an important part of the process by which creatures adapt and change within the limits of created kinds.2 Yet this has been controversial as well. I have argued that creationists have been far too quick to accept natural selection as an explanation when it has not actually been demonstrated (Lightner 2010a). Guliuzza has gone further equating natural selection with phantasm (Guliuzza 2012). With this kind of disagreement, how can we hope to understand the role of natural selection in our world?
Magical Descriptions and Powers
Part of the controversy is related to how natural selection is often portrayed, even in much of the scientific literature. Anthropomorphic terms are often applied to it. Clearly, nature has no mind and cannot select. The use of active verbs such as selects, preserves, favors, etc. are poetic, but not scientific ways of describing reality. So the term itself is an oxymoron. A farmer can select, for he has a mind. Nature cannot select, for it has no mind. This apparent “ghost in the machine” is one reason some evolutionists have rejected natural selection entirely (Fodor and Piattelli-Palmarini 2010).
A second problem is that some biologists speak as if natural selection can cause a new trait to appear. For example, they will claim that a trait arose due to selection pressure. This type of description was criticized by Endler, a renowned evolutionary biologist who studied vertebrate adaptation (Endler 1986). He pointed out what many creationists have since: that natural selection can only account for the change in frequency of a trait within a population; it does not explain how that trait originated (Catchpoole 2011). This is no small misunderstanding as former atheistic evolutionist Dr. David Catchpoole describes it as an eye-opening experience to him when someone clearly pointed out that natural selection cannot account for the origin of a trait. It was a pivotal point for him in his life—suddenly he was able to believe the Bible, even from the very first verse, and so become a biblical creationist (Catchpoole pers. comm.).
There are certainly plenty of other problems and associated details in how natural selection is often portrayed. To explore them all could fill volumes, but that is not the main point of this discussion. As will be seen, natural selection can be defined in a scientifically rigorous manner and modeled mathematically. The focus of the rest of the article is to survey fundamental concepts important to understanding the role natural selection may play in our world. More specifically, its role in speciation and adaptation will be considered, especially as it relates to mammals and birds.
A Scientific Syllogism
The Modern Synthesis (aka neo-Darwinism) essentially merged Darwin’s idea of natural selection with Mendelian genetics. It became well established over half a century ago. John Endler, a prominent evolutionary biologist, attempted to address misuse of the term natural selection and promote a logical, scientific basis for identifying it in his famous 1986 bookNatural Selection in the Wild. In this book he presents natural selection as a syllogism:
IF a population has:
- variation in traits with
- fitness differences (i.e., a consistent relationship between the trait and the ability to survive and reproduce, aka. differential reproduction), and
- heritability (implying that the trait is genetic)
Then the trait’s distribution in the offspring will be predictably different from that of the parental generation (if the population is not in equilibrium). (Lightner 2010a)
There are a few other points Endler makes in his book that are worth discussing here. To Endler, natural selection and evolution are not synonymous. Evolution must also account for the origin of the trait. Natural selection can only account for a change in the prevalence of a trait that already exists in a population. He criticized the definition of evolution used by population geneticists: that evolution is a change in allele frequency over time.
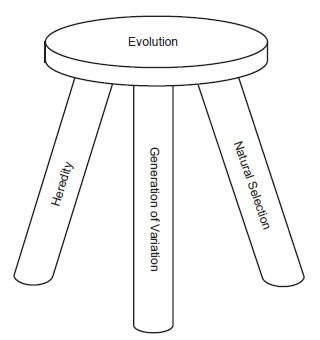
Fig. 1. Evolution’s three legged stool.
Using Endler’s definition, then, evolution can be understood like a stool (fig. 1) that is supported by three legs: the generation of variation, heredity, and natural selection (which occurs due to fitness differences). This basic idea has been echoed by other authors as well (Kirschner and Gerhart 2005).
Is accounting for the origin of variation necessary in a creationary paradigm? Since each kind was created separately there were certainly many traits placed in creatures at the beginning by their Creator. However, the creationary paradigm includes a severe genetic bottleneck in land creatures and birds at the time of the Flood. This places clear limitations on the amount of diversity that could have been present in those populations at the time. Examination of within kind variability in mammals (Lightner 2009) and birds (Lightner 2010b) confirms that creationists also must account for a significant amount of variation that has arisen since the time of the Flood. Some of this variation is clearly adaptive (Lightner 2014a). Will natural selection help explain how it became prominent in a specific population?
New Beneficial Mutations—Lost
If natural selection is considered deterministically, any rare beneficial mutation should eventually become fixed in the population. This is probably why natural selection often seems such a compelling explanation for adaptation. Evolutionists have long recognized that the frequency of a particular lineage will fluctuate over time. The fluctuations are called genetic drift and are treated stochastically in mathematical population genetics models (of those which take it into account). The reality is that the vast majority of beneficial mutations that arise should be lost due to genetic drift. The probability of loss decreases some if the population is small and growing, and generally increases if the population size is declining (Patwa and Wahl 2008).
This has profound implications for both evolutionary and creationary models. A beneficial mutation would need to repeatedly be generated before it would be expected to escape the effects of genetic drift and have a chance to become fixed in a population. The chance of a new beneficial mutation remaining in a population is improved a little if the population is small and growing, but a small population is the most unlikely place for a new beneficial mutation to occur if the underlying mechanism is random mutation. Instead, a large population is more likely to have enough individuals so random mutation might produce a beneficial allele—the very place where there is a higher probability of it being lost.
Fixing Alleles
Of course, not all mutations are lost. Further, there are clearly times where variation already present within a population (also known as standing variation) can change over time. There is an impressive body of literature in the field of mathematical population genetics that models how this may occur. One major focus has been to determine the probability that a beneficial allele will reach fixation, that is, the condition where it is found in every member of the population. This is an essential issue because it relates to speciation and adaptation. For example, it is common for there to be systematic differences in the alleles carried by different species—indicating a difference in which alleles have become fixed in those populations (Lightner 2014b).
Early on evolutionists became aware that adaptation by natural selection is more challenging to explain in vertebrates. In 1957 J. B. S. Haldane, a founder in the field of population genetics, published an article titled “The cost of natural selection” (Haldane 1957). Basically, animals such as mammals and birds have much longer generation times and smaller population sizes than bacteria or insects. There is simply not enough time, even in the evolutionary timescale, for natural selection to fix alleles within these populations (Batten 2005).
To resolve this dilemma, Kimura proposed the neutral theory of molecular evolution. He suggested that most of the genome has no function, so it is free to change by genetic drift. It seemed his evolutionary predictions were validated when it was found that only a small portion of the human genome encodes protein-coding genes, so “junk DNA” became a central theme of arguments supporting evolution. However, a steady stream of research has demonstrated that “junk DNA” really encodes a variety of critical regulatory RNAs (Faulkner et al. 2009; Tomkins 2013a, 2013b). The erosion of neutral theory is a major blow for evolutionists. They have no mathematically plausible way to explain patterns such as the millions of differences between human and chimp DNA which supposedly arose during the 100,000 to 300,000 generations since their purported divergence (Carter 2009).
Creationist John Sanford, one of the developers of the population genetics program Mendel’s Accountant, has publicized some of the implications of both natural selection and genetic drift. Given that most mutations are near neutral, natural selection has little effect on them. Most beneficial mutations are not effectively fixed, nor are most deleterious mutations effectively removed. The majority of the changes in allele frequencies are the result of genetic drift. In fact, the small proportion of beneficial mutations that survived removal by genetic drift (less than 1%) and eventually became fixed, were usually fixed by genetic drift rather than natural selection (Rupe and Sanford 2013).
Further problems occur when multiple beneficial mutations are present at the same time. Competition between two beneficial mutations can reduce the probability of the second one being fixed. In asexual organisms this is termed clonal interference (Patwa and Wahl 2008, pp. 1284–1286). It affects eukaryotic organisms as well. An attempt was made to quantify this in a recent numerical simulation study (Sanford, Baumgardner, and Brewer 2013). This has profound implications for the role natural selection may play in adaptation; adaptation is often the result of changes in a suite of traits involving a number of genes (Dong et al. 2014; Tekola-Ayele et al. 2014).3 If there are problems accounting for how natural selection can effectively make changes in one allele at a time, how is it expected to account for adjustments in polygenic networks that involve epistatic interactions? A recent review on adaptation to high altitude suggests a lack of plausibility for natural selection being a major contributing mechanism (Lightner 2014a).
There is considerably more that has been discussed in the population genetics literature, most of which is far beyond the scope of this discussion (Patwa and Wahl 2008). Many different parameters can be varied in the calculations. This can include accounting for both beneficial and deleterious mutations, interactions between mutations, and other variables. There is excellent evidence that population size fluctuates in mammals and birds; strength of selection can vary as well (Grant and Grant 2014). Several studies have taken these two variables into account (Wahl 2011). One of these suggests that fluctuating population size will decrease the effect of genetic drift (Waxman 2011). There has also been more published by creationists in the field of population genetics, including papers presented at the Biological Information: New Perspectives Symposium held at Cornell University in 2011 (Marks et al. 2013). In the end, the models suggest that natural selection cannot possibly play the prominent role in adaptation that the compelling stories we are told suggest.
Comparison to Examples of Artificial Selection
Darwin compared what was known from animal breeding to his ideas about natural selection. It might be helpful to do so here as well. A dairy farmer who wants to increase milk production in his herd has some powerful tools at his disposal. The dairy industry has been a leader in record keeping. The farmer can breed his cows with semen from bulls that had been extensively tested and shown to sire daughters with exceptional milk production. This allows him to have far more power to change the genetics of his herd than would be available in a natural population since he can bring in genetic material from outside the herd to address a specific need. If he focuses on just one trait, some impressive, rapid progress can be made.
The problem is that many traits are interrelated. A gallon of milk weighs about 8½ lb (3.9 kg). If a cow produces more milk, she needs strong udder attachments to support the extra weight. Otherwise the suspensory system may give out and her teats may drag on the ground. This increases her risk of disease (for example, mastitis, which decreases milk production and can render the milk she does produce unsalable) and injury (which can further increase the risk of mastitis). Additionally, carrying more milk increases stress on the feet and legs, which can result in lameness. Fortunately, the record keeping allows the farmer to select for more than one trait. However, as the number of traits a farmer wishes to select for increases, the rate of progress slows.
Several other factors influence progress towards a goal. Generally, initial progress is more rapid since there is a larger difference between production in his herd and the average production of the daughters of the sire he chooses. Progress is also influenced by how highly heritable the trait is. In some cases, selecting for one trait (for example, milk production) can be associated with a loss in another area (for example, fertility). Sustained intentional selection within the U.S. dairy industry over the past half century has been associated with an impressive increase in milk production along with improved soundness in udders, feet, and legs. In contrast, fertility has not enjoyed such an improvement (Cassell 2009). Another factor that may not be obvious to those outside the industry is that improvement in management has played a role in realizing these gains. This amounts to the farmer changing the environment for the cow in order to enjoy the benefits of his breeding program.
The other side of the coin is seen when trying to eliminate genetic diseases. Spider lamb syndrome is a semi-lethal recessive disorder that showed up in black-faced sheep in the mid 1970s. The problem was that to eliminate this disease from the herd, a farmer needed good records to identify the carriers. Depending on the management system used, this could sometimes be challenging. Eventually, a genetic test was developed to enable identification of carriers.4
Artificial selection is powerful, but anyone who has been involved in this area soon recognizes that it has obvious limitations. Good records are essential and only a limited number of traits can be selected for at once if real progress is to be made. Sometimes selecting for one trait has a negative effect on a different desired trait. While humans can select against normal-appearing carriers of a recessive disease given the proper tools (good records or genetic testing), natural selection cannot. Natural selection will have no effect until homozygous individuals appear; it is powerless to eliminate recessive deleterious mutations. The population genetics models and examples of selection in agriculture are helpful in discerning between a realistic appraisal of natural selection and the “magical story-telling” which abounds in many presentations.
Genetic Patterns and Statistical Tests
Despite the dismal predictions about the importance of natural selection in vertebrates based on population genetics models and the limitations recognized from animal breeding, many biologists in the field are convinced that there is excellent evidence that natural selection has occurred and that it is important in accounting for adaptation. In addition to significant morphologically based studies, many of which Endler highlights in his book, genetic-based studies often come to the same conclusion. Why this apparent discrepancy?
It needs to be remembered that whenever math is applied to scientific questions there are always assumptions. This is true of population genetics models briefly mentioned above (for example, about population size, mutation rate, selection strength, interaction between multiple mutations, etc.). It is equally true of statistical tests intended to identify if natural selection accounts for observed patterns. Statistical tests are helpful in identifying non-random patterns, but whether the pattern is best understood as being a result of selection is a different matter. For example, it is known that demographic events, such as migration, expansions, and bottlenecks, can result in similar non-random patterns (Vitti, Grossman, and Sabeti 2013). Despite this, many articles are written as if selection is the definitive cause of the non-random pattern (for example, Ren et al. 2014).
There are additional factors that need to be considered as well. For example, one helpful review of statistical tests gave an example of the HARIF gene, which expresses non-coding RNA during brain development (Vitti, Grossman, and Sabeti 2013). It is described as highly conserved between chimps and other vertebrates, but humans had 40 times more substitutions than would be expected had the locus neutrally evolved. The problem here is obvious. The statistical test certainly has picked up on an obvious difference in the sequence of this region in humans compared to other vertebrates. However, the interpretation that the difference is a result of selection is ill-founded since humans do not share common ancestry with other vertebrates.
From a creationary perspective, then, comparisons need to be made within a created kind to reasonably conclude that selection accounts for the pattern. Of course there are many examples where this is done. In general mutations are assumed to occur randomly, though a few models account for the fact that transitions are more common than transversions; some models consider recombination. In regions where little variability is observed, it is generally interpreted as evidence of purifying selection. Is this the case, or might there be cellular mechanisms suppressing mutation in the region, or enhancing it in others?
A specific example may help in clarifying the problem. The MC1R gene is highly polymorphic in humans with over 100 alleles with non-synonymous (that is, affecting the amino acid) changes having been identified to date (Dessinoti et al. 2011). Some of these alleles are associated with red hair and increased susceptibility to skin cancers. MC1R variants are common in European populations and some biologists assume that this locus is under selection. A study published in 2000 used statistical tests which suggested that there was no selection occurring in populations outside Africa, but strong purifying selection effectively eliminated all non-synonymous mutations within the African populations they sampled. The authors argued that this made sense because amino acid sequence changes have been documented to affect function and some can do so even when a single allele is present (that is, some alleles have semidominant expression rather than recessive) (Harding et al. 2000).
Superficially this may sound reasonable, but in fact this is completely inconsistent with what is known about this well characterized gene. Amino acid variants vary widely in how strongly they are associated with cancer; in many no such association has been demonstrated. Even of those variants most strongly associated with the disease, most individuals carrying them never develop skin cancer. Further, if they do develop cancer, the average age of onset is over 50 years old—far beyond the prime childbearing years (Lachiewicz et al. 2008). It is pure fantasy to think that the absence of amino acid variants among those Africans sampled in the study is due to natural selection. Yes, amino acid variants may well be harmful to inhabitants of that region as the authors of the study pointed out. However, natural selection is powerless to prevent the transmission of such variants to the next generation. A more plausible reason for this pattern is that there are designed mechanisms which influence the pattern of mutation in this gene in response to environmental stimuli.
Some statistical comparisons involve comparing the ratio of non-synonymous mutations (those that change the amino acid sequence of the protein) to synonymous mutations (those that do not change the amino acid). The assumption is that synonymous mutations are neutral and provide a measure of underlying mutation rate. The problem is that mounting evidence suggests that non-synonymous mutations are not actually neutral. Evidence suggests that codon usage affects the rate of protein synthesis, and regulating the rate of protein synthesis is essential to insure proper folding of the protein (D’Onofrio and Abel 2014). Another review, which focused on statistical tests as a means to differentiate between neutral models (where allelic patterns are due to drift) and Darwinian selection, brings out other limitations and assumptions involved in various statistical tests (Nielsen 2005). In the discussion of positive selection, the author briefly mentions examples of where segregation distortion (also known as meiotic drive) appears to be the underlying cause. This means that traits are not being passed on to the next generation as one would expect if Mendelian inheritance is involved. It is important to recognize that this is not selection in the traditional sense that someone like Endler would view it. In fact, Endler’s syllogism assumes standard Mendelian inheritance.
Meiotic Drive: Another Means for Fixing Alleles
Meiotic drive was described in the literature well over half a century ago (Sandler and Novitski 1957). It can be defined as an alteration in the process of meiosis that results in a heterozygote (organisms carrying two different alleles for a gene) preferentially transmitting one allele over the other. Some use the term more broadly to refer to other non-Mendelian transmission even when the timing of the cause is not during meiosis. A number of mechanisms have been proposed and/or described over the years (Harvey et al., 2014; Montchamp-Moreau 2006; Zimmering, Sandler, and Nicoletti 1970).
Biased gene conversion is one mechanism resulting in meiotic drive. During meiosis chromosomes pair up and crossing over occurs. Gene conversion also occurs at this time and begins in a similar way as crossing over, with an enzyme-induced double stranded break in the DNA. However, with gene conversion the sequence on one strand is copied over onto the other. There are times where the cut occurs more frequently on one strand, and the sequence from the other strand is preferentially transmitted; this is known as biased gene conversion. Biased gene conversion can result in fixation of one allele even in the absence of natural selection or drift.5
Consistent with the naturalistic bias of evolutionary scientists, biased gene conversion has been assumed to be random with respect to fitness. It is recognized that generally it results in an increased transmission of alleles with GC over AT. It has been proposed that biased gene conversion can be distinguished from natural selection because it should not be consistently correlated with adaptation, but natural selection would be (Galtier and Duret 2007). What if this is not true? Living things are amazingly designed with the capacity to control gene expression, RNA editing, and many other processes. Given that gene conversion is a normal part of meiosis, why wouldn’t it have been designed to enable the amplification of adaptive alleles in a population?
Here, then, is an opportunity to compare what can be considered reasonable predictions of the evolutionary and creationary models. The evolutionary model assumes universal common ancestry and that changes occurring in organisms throughout natural history are explainable by natural processes. In contrast, the biblical creation model recognizes that creatures were created according to their kinds (limited common ancestry) to reproduce and fill the earth. If the latter is true, then mechanisms such as biased gene conversion and other forms of meiotic drive are likely to account for a significant amount of the change in gene frequency necessary for adapting to various habitats around the world. So I predict that biased gene conversion will be found to increase the frequency of adaptive alleles in populations.
If the genome has DNA-editing abilities resulting in a bias for adaptive mutations and/or mechanisms to increase the frequency of existing adaptive alleles, then this explains much of the discrepancy between the conclusions of biologists using a statistical test to identify selection and the predictions of population genetic models.6 The non-random patterns identified by the test are often caused by designed mechanisms that enable creatures to adapt, yet credit has been given to natural selection because such designed mechanisms were assumed to not exist. This would be another classic case of where the neo-Darwinian worldview hindered an understanding of the world around us.
A Case Study: Darwin’s Finches
Certainly not all examples of natural selection are merely inferred from statistical tests. At times additional information is collected allowing one to make a more compelling case that natural selection has occurred. Probably the most–well-known example involves the impressive work done by Peter and Rosemary Grant with finches on the small island of Daphne Major in the Galápagos. They primarily focused on the medium ground finch (Geospiza fortis), but they also collected some data on other finches as well. However, the many details they provide from their 40 years of study indicate that natural selection is not operating in the way it is commonly assumed to operate (Grant and Grant 2014).
There were a number of advantages in choosing these island finches for study. The population was small enough so individuals could be measured and then tagged for easy recognition. Beak measurements (length, width, and depth) and body size (wing and tarsus length) were recorded. Breeding was tracked and heritability was determined. Beak dimensions were variable, determined to be highly heritable7, and observed to be correlated with diet. Birds with larger beaks were able to crack and consume the larger and harder seeds. Those with smaller beaks were dependent on smaller and softer seeds.
Rather than detect constant natural selection fitting the finches to the environment, natural selection only appeared to be operating at discrete times during the study when the environmental conditions became unusually harsh. For example, after a drought in 1977, only one very large nestling and 15% of the adults remained. There was a size-biased mortality in the birds. Many smaller birds were found dead and many more of similar size were missing and presumed dead.9 Given that the small and soft seeds these birds feed on were depleted early during the drought, it is not surprising the birds dependent on them were wiped out. Obviously, the average beak size of the population became larger, which qualified as evolution as typically defined by population geneticists.
Prior to another severe drought in 1985, extremely wet years had increased the availability of small and soft seeds considerably. During this drought, it was the large and hard seeds that became depleted and the average beak size shifted in the opposite direction. In other years mathematically it appeared that there was selection for a trait, but average beak size did not change as predicted because one or more linked traits were selected in a different direction. Of course, sometimes a linked trait can change even if it is not the one being selected.
By definition, natural selection cannot occur unless there is variability in a trait. There was certainly variation in beak size when their study began. It was useful as the birds exploited the variety of seeds present on the island. When natural selection was clearly documented, it eliminated much of the useful variety, and oscillated in direction. This is not the pattern suggested by Darwin, and could easily eliminate the birds altogether. What allowed them to survive?
One factor that provided long term stability was low levels of hybridization that occurred between the medium ground finch and two other species of finch on the island. It had the effect of reintroducing some of the lost variability so the population could better exploit the variety of seed sources available again after the rains returned. So in this example natural selection could be better thought of as catastrophic elimination. Especially because of its oscillating pattern, it was a threat to the survival of the birds. If we compare this to artificial selection, there is a capricious madman doing the selecting; he cannot make up his mind what he wants. Despite this, the destructive effects were overcome, in part by hybridization.
The oscillating pattern of natural selection described by the Grants is far more devastating to a naturalistic view of life than evolutionists seem to recognize. Progress in artificial selection is made by conscious, focused effort towards a goal. Historically, it was assumed natural selection would do the same if environmental conditions remained relatively constant in an area. It is now clear that years which deviate significantly from the average can have a dramatic negative impact on the population. These extreme conditions can eliminate traits which would normally be useful in that environment. In this case natural selection is a far more logical explanation for extinction than adaptation.
Summary
Hopefully this brief overview will help bring clarity to our thinking on natural selection, specifically as it relates to vertebrates such as mammals and birds. Natural selection is a valid phenomenon despite the fact that the phrase can be misleading and misused. However, multiple lines of evidence suggest that it has not played a major role in enabling mammals or birds to diversify and adapt to their environment.
What has happened is that statistical tests, which can be used to screen for natural selection, have generally not been followed up to validate that selection is actually the cause of the pattern. Demographic events such as migration, expansions, and bottlenecks have generally been ignored. Biased gene conversion and other forms of meiotic drive have been ignored or assumed to be random phenomena. This has resulted in conclusions about the importance of natural selection that are at odds with other lines of evidence. This has hindered scientific understanding of the true basis for adaptation. A reasonable prediction of the creation model is that mechanisms such as biased gene conversion were designed to aid in adjusting allele frequency in populations, enabling creatures to adapt.
References
Batten, D. 2005. Haldane’s dilemma has not been solved. TJ 19, no. 1: 20–21. http://creation.com/haldanes-dilemma-has-not-been-solved.
Carter, R. W. 2009. The slow, painful death of junk DNA. Journal of Creation 23, no. 3: 12–13. http://creation.com/junk-dna-slow-death.
Cassell, B. 2009. Sire evaluations for health and fitness traits. Virginia Cooperative Extension. Virginia Polytechnic Institute and State University, Blacksburg, Virginia. http://pubs.ext.vt.edu/404/404-087/404-087_pdf.pdf.
Catchpoole, D. 2011. Defining terms—John Endler’s refreshing clarity about ‘natural selection.’ Journal of Creation 25, no. 22: 19–21.
Dessinoti, C., C. Antoniou, A. Katsambas, and A. J. Stratigos. 2011. Melanocortin 1 receptor variants: Functional role and pigmentary associations. Photochemistry and Photobiology 87, no. 5: 978–987.
Dong, K., N. Yao, Y. Pu, X. He, Q. Zhao, Y. Luan, W. Guan, S. Rao, and Y. Ma. 2014. Genomic scan reveals loci under altitude adaptation in Tibetan and Dahe pigs. PLoS One 9, no. 10: e110520.
D’Onofrio, D. J., and D. L. Abel. 2014. Redundancy of the genetic code enables transcriptional pausing. Frontiers in Genetics 5: 140. doi: 10.3389/fgene.2014.00140.
Endler, J. A. 1986. Natural selection in the wild. Princeton University Press: Princeton, New Jersey.
Faulkner, G. J., Y. Kimura, C. O. Daub, S. Wani, C. Plessy, K. M. Pirvine, K. Schroder, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nature Genetics 41, no. 5: 563–571.
Fodor, J., and M. Piattelli-Palmarini. 2010. What Darwin got wrong. New York, New York: Farrar, Straus, and Giroux.
Galtier, N., and L. Duret. 2007. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends in Genetics 23, no. 6: 273–277.
Grant, P. R., and B. R. Grant. 2014. 40 years of evolution: Darwin’s finches on Daphne Major Island. Princeton, New Jersey: Princeton University Press.
Grant, P.R., T. D. Price, and H. Snell. 1980. The exploration of Isla Daphne Minor. Noticias de Galápagos 31: 22–27. http://www.darwinfoundation.org/datazone/media/pdf/31/NG_31_1980_Grant_etal_Exploration.pdf.
Guliuzza, R. J. 2012. Darwin’s sacred imposter: Answering questions about the fallacy of natural selection. Acts & Facts 41, no. 2: 12–15. http://www.icr.org/article/6590/.
Haldane, J. B. S. 1957. The cost of natural selection. Journal of Genetics 55: 511–524.
Harding, R. M., E. Healy, A. J. Ray, N. S. Ellis, N. Flanagan, C. Todd, C. Dixon, et al. 2000. Evidence for variable selective pressures at MC1R. American Journal of Human Genetics 66, no. 4: 1351–1361.
Harvey, A. M., D. G. Rehard, K. M. Groskreutz, D. R. Kuntz, K. J. Sharp, P. K. Shiu, and T. M. Hammond. 2014. A critical component of meiotic drive in neurospora is located near a chromosome rearrangement. Genetics 197, no. 4: 1165–1174.
Kirschner, M. W., and J. C. Gerhart. 2005. The plausibility of life: Resolving Darwin’s dilemma. New Haven, Connecticut: Yale University Press.
Lachiewicz, A. M., M. Berwick, C. L. Wiggins, and N. E. Thomas. 2008. Epidemiologic support for melanoma heterogeneity using the surveillance, epidemiology, and end results program. Journal of Investigative Dermatology 128, no. 5: 1340–1342.
Lightner, J. K. 2009. Karyotypic and allelic diversity within the canid baramin (Canidae). Journal of Creation 23, no. 1: 94–98.
Lightner, J. K. 2010a. Matters of fact . . . What is natural selection? Creation Matters 15, no. 4: 10–11. https: //creationresearch.org/index.php/extensions/s5-creation-matters/cm-archive?task=document.viewdoc&id=670.
Lightner, J. K. 2010b. Identification of a large sparrow-finch monobaramin in perching birds (Aves: Passeriformes). Journal of Creation 24, no. 3: 117–121.
Lightner, J. K. 2013. Meiotic recombination—designed for inducing genomic change. Journal of Creation 27, no. 1: 7–10.
Lightner, J. K. 2014a. Adaptation of endotherms to high altitudes. Creation Research Society Quarterly 50, no. 3: 132–140.
Lightner, J. K. 2014b. Developmental system plasticity—A brief initial assessment of extent, design, and purpose within the creation model.Journal of Creation 28, no. 3: 67–72.
Marks, R. J. II, M. J. Behe, W. A. Dembski, B. L. Gordon, and J. C. Sanford. eds. 2013. Biological information—New perspectives. Singapore, Asia: World Scientific Publishing Co.
Montchamp-Moreau, C. 2006. Sex-ratio meiotic drive in Drosophila simulans: cellular mechanism, candidate genes and evolution. Biochemical Society Transactions 34 (pt 4): 562–565.
Nielsen, R. 2005. Molecular signatures of natural selection. Annual Review of Genetics 39: 197–218.
Patwa, Z., and L. M. Wahl. 2008. The fixation probability of beneficial mutations. Journal of the Royal Society Interface 5, no. 28: 1279–1289.
Purdom, G. 2007. Is natural selection the same thing as evolution? In The New Answers Book, ed. K. Ham, 271–282. Green Forest, Arkansas: Master Books.
Ren, L., X. J. Tan, Y. F. Xiong, K. Xu, Y. Zhou, H. Zhong, Y. Liu, Y. H. Hong, and S. J. Liu. 2014. Transcriptome analysis reveals positive selection on the divergent between topmouth culter and zebrafish. Gene 552, no. 2: 265–271.
Rupe, C. L., and J. C. Sanford. 2013. Using numerical simulation to better understand fixation rates, and establishment of a new principle: Haldane’s ratchet. In Proceedings of the Seventh International Conference on Creationism, ed. M. Horstemeyer. Pittsburgh, Pennsylvania: Creation Science Fellowship.
Sandler, L., and E. Novitski. 1957. Meiotic drive as an evolutionary force. American Naturalist 91, no. 857: 105–110.
Sanford, J. C., J. R. Baumgardner, and W. H. Brewer. 2013. Selection threshold severely constrains capture of beneficial mutations. In Biological Information—New Perspectives, ed. R. J. Marks II, M. J. Behe, W. A. Dembski, B. L. Gordon, and J. C. Sanford, 264–297. Singapore, Asia: World Scientific Publishing Co.
Tekola-Ayele, F., A. Adeyemo, G. Chen, E. Hailu, A. Aseffa, G. Davey, M. J. Newport, and C. N. Rotimi. 2014. Novel genomic signals of recent selection in an Ethiopian population. European Journal of Human Genetics. doi: 10.1038/ejhg.2014.233.
Tomkins, J. P. 2013a. The Human Beta-Globin Pseudogene Is Non-Variable and Functional. Answers Research Journal 6: 293–301. https://answersingenesis.org/genetics/humangenome/the-human-beta-globin-pseudogene-is-non-variable-and-functional/.
Tomkins, J. P. 2013b. Alleged Human Chromosome 2 “Fusion Site” Encodes an Active DNA Binding Domain Inside a Complex and Highly Expressed Gene—Negating Fusion. Answers Research Journal 6: 367–375. https://answersingenesis.org/genetics/dna-similarities/alleged-human-chromosome-2-fusion-site-encodes-an-active-dna-binding-domain-inside-a-complex-and-hig/.
Vitti, J. J., S. R. Grossman, and P. C. Sabeti. 2013. Detecting natural selection in genomic data. Annual Review of Genetics 47: 97–120.
Wahl, L. M. 2011. Fixation when N and s vary: Classic approaches give elegant new results. Genetics 188, no. 4: 783–785.
Waxman, D. 2011. A unified treatment of the probability of fixation when population size and the strength of selection change over time. Genetics 188, no. 4: 907–913.
Wilkins, A. S. 2012. Book review: Evolution: A view from the 21st century by J. A. Shapiro. Genome Biology and Evolution 4, no. 4: 423–426.
Zimmering, S., L. Sandler, and B. Nicoletti. 1970. Mechanisms of meiotic drive. Annual Review of Genetics 4: 409–436.