The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
In recent years, a special focus of microbiology research has been on the certain groups of bacteria (such as the superphylum Planctomycetes-Verrucomicrobia-Chlamydiae—PVC bacteria) because they exhibit certain characteristics which are unusual for prokaryotes, and which are also shared by eukaryotes. Such characteristics include a nuclear membrane, budding reproduction, sterol biosynthesis, and condensed nucleoids. These characteristics challenge the long-held concept that the presence or absence of a nuclear membrane is enough to differentiate between prokaryotes and eukaryotes and also seemingly support the evolutionary idea of the transition from prokaryotes to eukaryotes due to seemingly similar structures shared by these two domains of life.
However, upon closer examination, many protein sequences which are involved in these structures and processes shared by prokaryotes and eukaryotes show low sequence homology and are similar in structure only. Therefore, these proteins can only be said to be analogous to each other, rather than homologous, which is required for evolutionary descent with modification.
Furthermore, PVC bacteria are not thought to be the direct ancestors of eukaryotes, despite their analogous cellular characteristics. Also, even though the α-proteobacteria are thought to be the ancestor of the eukaryotes’ mitochondria, its energetic capabilities are questionable, which are hypothesized to be necessary for the expansion of eukaryotic cell complexity, compared to those of other bacteria, such as that of species in the genus Rhodobacter. Some bacteria have also been discovered that contain energy-producing compartments called the anammoxosome, which contradicts the endosymbiotic theory, which states that energy-producing bacteria became mitochondria, which was necessary to induce eukaryotic cellular complexity.
Introduction
According to the well-known paradigm, eukaryotes are defined by the presence of a nuclear membrane surrounding their DNA, which is wound up into several pairs of chromosomes, as well as the presence of endocytosis and an endomembrane system, thereby compartmentalizing different cellular processes and separating them from one another inside the cell (de Duve 2007). This molecular characteristic separates them from prokaryotes whose genome is much smaller, and condensed into a single, circular DNA moleculep. The transition from prokaryotes to eukaryotes (PET) counts as a major obstacle for evolutionary theory between these two types of cells (Maynard Smith and Szathmáry 1995).
Until now there have been two basic theories on the evolution of cellular complexity. According to one theory, cellular complexity came about via endocytosis between bacteria and archaea. The problem with this theory is that, in practice, newly made constituents are always inserted into pre-existing membranes (omnis membrane e membrana), which subsequently divide (de Duve 2007). Membrane fusion between archaea and bacteria has never been observed. According to another theory, membrane invagination occurred after the digestion of engulfed material by the cell (Lonhienne et al. 2010). The invaginated membrane grew more and more convoluted, and then more specialized into different compartments, such as the endoplasmic reticulum (ER), Golgi apparatus and lysosomes (de Duve 2007).
However, the view that the presence of a nuclear membrane separating prokaryotes and eukaryotes from one another has been challenged in past years by the discovery of a number of different bacterial species in the superphylum Planctomycetes-Verrucomicrobia-Chlamydiae (PVC bacteria). Such species include the model organism Gemmata obscuriglobus, Chthoniobacter flavus, Pedosphaera parvula, Verrucomicrobium spinosum, and Prosthecobacter dejongeii. These species also have certain membrane coat proteins (MC proteins), such as clathrin, Sec31, α-and β’-COP, and nucleoporin (Forterre and Gribaldo 2010; Santarella-Mellwig et al. 2010), which are analogous to those found in eukaryotes. MC proteins are involved in vesicular transport of macromolecules between different compartments in the eukaryotic cell, such as between the Golgi-apparatus and the ER (Bonifacino and Lippincott-Schwartz 2003). These evidences would support the invagination-hypothesis mentioned previously.
On a cellular level, G. obscuriglobus has a cell membrane, covered by an external cell wall. Just underneath the cell membrane is an intracellular membrane, which partially peels away from the outer cell membrane at one pole of the cell, and forms a stalk, which is connected to an inner membrane surrounding the condensed nucleoid. The part of the cell which separates from the outer cell membrane forms a compartment called the paryphoplasm. No ribosomes are found here, only in the neighboring compartment called the riboplasm and also near the nucleoid. Proteins can be taken up externally and enter the paryphoplasm (Fuerst and Sagulenko 2014). A diagram of G. obscuriglobus can be seen in Fig. 1. A description of eukaryote-like proteins in G. obscuriglobus can be seen in Table 1 (Fuerst and Sagulenko 2012). The phylum planctomycetes, which G. obscuriglobus belongs to, also exhibits unusual properties for prokaryotes, such as budding reproduction, sterol biosynthesis (which is also characteristic of eukaryotes), and permanently condensed nucleoids (Lonhienne et al. 2010). The enzymes squalene monooxygenase (SQMO) and oxidosqualene cyclase (OSC) are responsible for the initial steps in the biosynthesis of sterol, which is a component of the eukaryotic membrane. BLAST searches at a maximum E-value of 10-8 showed that these two genes have homologs in G. obscuriglobus and other species (Pearson, Budin, and Brocks 2003). In G. obscuriglobus (as well as α-proteobacteria), cell division occurs via budding at one point of the perimeter of the mother cell until completion, with nucleoid membranes forming from the intracytoplasmic membrane (ICM). The bacterial cell division protein FtsZ is missing in planctomycetes (Lee, Webb, and Fuerst 2009).
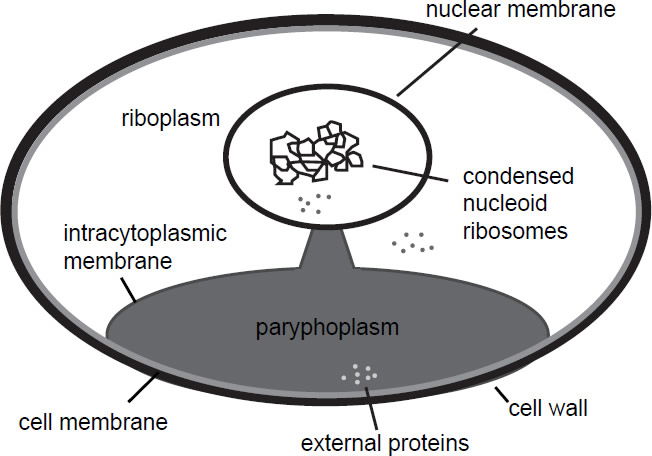
Fig. 1. Schematic depiction of cellular structure of Gemmata obscuriglobus. Like eukaryotic cells, this species has a cell wall, a nuclear membrane, condensed DNA, and is compartmentalized. External proteins enter a special compartment called the paryphoplasm.
| Gene/Protein Domain | Function |
|---|---|
| cadherin domain | N-terminal export signal |
| calx-β domain | N-terminal export signal |
| clathrin | Membrane coating, vesicle traffic |
| cytochrome domains | Energy production |
| discoidin domains | Cell adhesion |
| integrin | Part of cytoskeleton |
| Sec31 | Membrane coating, vesicle traffic |
| thrombospondin domain | N-terminal export signal |
| α-COP protein | Membrane coating, vesicle traffic |
| β’-COP protein | Membrane coating, vesicle traffic |
G. obscuriglobus exhibits mixed characteristics of both prokaryotes and eukaryotes, thus challenging the long-held views of the sharp distinction between these two domains of life, suggesting that a number of molecular “missing links” have been found which connect prokaryotes with eukaryotes, thereby making the transition more fluid. Despite all of these considerations however, evolutionary theory encounters three serious roadblocks.
Materials and Methods
The data for Fig. 2 was taken from the supplementary data from Fuchsman and Rocap (2006). The figure was generated in R, version 3.3.1.
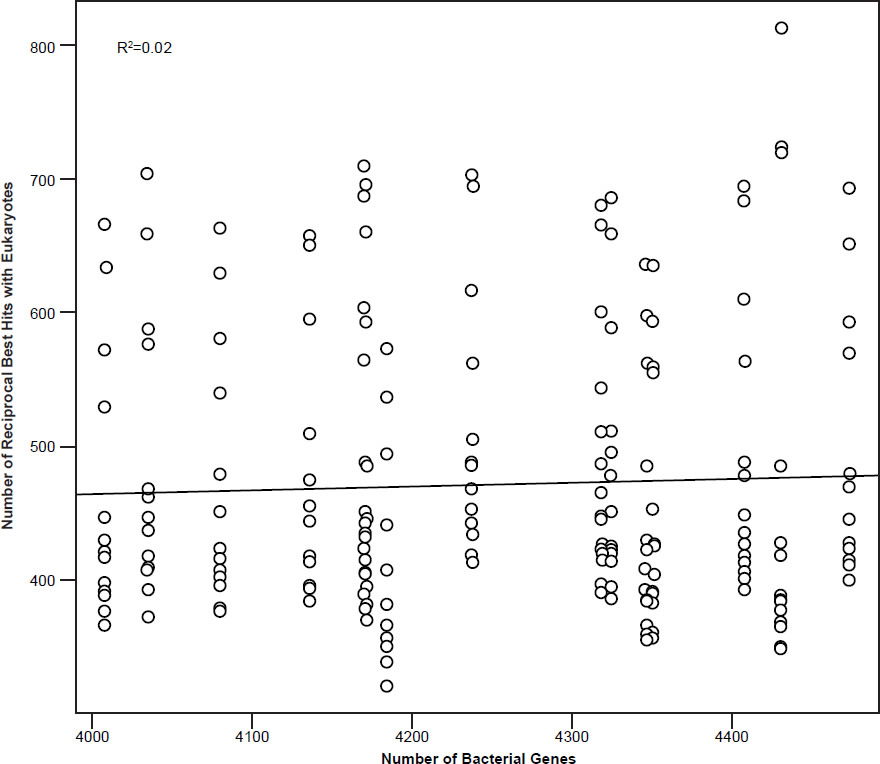
Fig. 2. Number of reciprocal BLAST hits to eukaryotic genes according to the number of genes in 33 bacterial genomes (cutoff E-value: 10-10). Data taken from supplementary data from Fuchsman and Rocap (2006). R2 = 0.02.
Prokaryotic proteins differ from eukaryotic ones
One of the major problems that the PET theory has is that the organization and characteristics of prokaryotic and eukaryotic genes are quite different. One would expect that during PET the gene content would gradually undergo a steady turnover (Staley, Bouzek, and Jenkins 2005). Introns would also have to be introduced into prokaryotic genes. Yet several papers have shown that only a small number of eukaryotic signature proteins have been detected in bacterial species such as P. dejongeii, Gemmata sp. WA-1, and Kuenenia stuttgartiensis (Wagner and Horn 2006). Therefore, these genes are thought to be the result of lateral gene transfer and not via descent. For example, the distribution of homologs of metazoan α2-macroglobulins occurs in multiple different bacterial clades, suggestive of LGT (Budd et al. 2004). According to a study by Fuchsman and Rocap (2006), which studied the number of reciprocal BLAST hits (cutoff E-value: 10-10) between 18 archaeal, 134 bacterial, and 14 eukaryotic genes showed that the number of reciprocal hits increased steadily until it reached 4000 host bacterial genes, but then plateaued. Indeed, when comparing reciprocal BLAST hits between 33 bacterial species and the 14 eukaryotic ones from the Fuchsman and Rocap study, which had more than 4000 genes in their genomes, the correlation between the number of bacterial genes and the number of hits was only 0.138 (insignificant), whereas the R2 value was also only 0.02 (fig. 2). What this means is that after a certain limit there are actually less reciprocal best hits with eukaryotic genes than expected. This is evidence which is in contradiction to the PET gene turnover process.
These analyses used fairly high E-values, in order to find all possible homologs, which might even have led to false positives. However, studies by Devos et al. (2004) and Santarella-Mellwig et al. (2010) concluded that no prokaryotic MC proteins were detected by sequence similarity searches; rather, prokaryotic and eukaryotic MC proteins are thought to be similar only structurally. Santarella-Mellwig based these conclusions on their analysis of 687,835 eubacterial proteins in 162 complete and 13 incomplete proteomes, 60,382 archebacterial proteins from 27 complete proteomes, and 231,229 eukaryotic proteins in 23 complete proteomes. Furthermore, they found that the GC content of prokaryotic MC-like genes are not significantly different from those of other planctomycete genes, and not significantly similar to eukaryotic proteins. Therefore, these authors conclude that both eukaryotic and prokaryotic MC proteins evolved separately, by convergent molecular evolution.
Some evolutionists may argue that prokaryotic and eukaryotic MC proteins underwent divergent evolution from common ancestor genes. They claim that during long periods of evolution, even though sequence similarity might be lost, structural similarity is retained (Devos and Reynaud 2010). However, it is hard to imagine that with a high degree of sequence turnover, affecting all parts of the protein, the protein would not lose its structurally determined function. Furthermore, this is only speculation, since neither the original proteins, nor the process of sequence erosion was ever observed.
Contradictions as to which bacterial group is the transition towards eukaryotes
The second major obstacle for the PET theory is that the PVC superphylum might not be the group of species among which the actual PET occurred. As to what kind of species was basal to LECA (the Last Eukaryotic Common Ancestor), there is only much hypothetical speculation which does not name any concrete species. According to some theories, the eukaryotes originated from archaea (Koonin 2015). According to another hypothesis, MC proteins were present even in LUCA (the Last Universal Common Ancestor), whereby they would have been subsequently retained only in Eukarya and PVC bacteria. According to yet another hypothesis, MC proteins appeared in PVC bacteria during PET (Forterre and Gribaldo 2010). However, 16S rRNA gene trees have differed significantly from one another as to the position of Planctomyces, with some trees placing them even at the root of Bacteria (Brochier and Philippe 2002; Fuchsman and Rocap 2006). Instead, other evolutionary studies conclude that eukaryotes are linked more to archaebacteria and proteobacteria, and alga and plants to cyanobacteria (McInerney et al. 2011). Indeed, even the individual phyla pertaining to the PVC superphylum have no obvious similarities to one another (Wagner and Horn 2006), meaning that they belong to different apobaramins.
Mere analogy between organelles is not enough for PET
The third obstacle for PET is that bacterial eukaryote-like structures are merely analogous and not homologous. Homology, or sequence similarity of gene sequences between two groups is supposed to be evidence for their relatedness (Bergman 2001). On the other hand, mere analogy means the existence of genes or proteins in two separate species which fulfill the same function yet are not sequentially similar. Analogy by itself is not enough for PET.
For example, the presence of a nuclear membrane-like structure as well as an analogous vesicular endocytosis system in some species of bacteria speaks of a certain level of cellular complexity for these kinds of organisms. Some evolutionists claim that these characteristics must have been present early on during the cellular stage of evolution in LUCA (Forterre 2011). Forterre and Gribaldo (2010) state that “we should definitely stop thinking of bacteria in terms of simple ‘lower’ organisms”. These are rather telling admissions about complexity supposedly appearing rapidly early on in evolution, which are not isolated phenomena. For example, cnidarians such as jellyfish also show relatively complex cell and tissue types, even though they are fairly close to the base of the evolutionary tree in Animalia (O’Micks 2015).
The membrane and compartment system
Classically, the greatest difference between prokaryotes and eukaryotes is the nuclear membrane. Also important is the presence of the nuclear pore complex (NCP) within the nuclear membrane, which regulates the exchange of molecules between the nucleoplasm and the cytoplasm.
For example, in G. obscuriglobus, the nuclear membrane analog is an extension of the cell membrane and not that of the endoplasmatic reticulum (ER) as in eukaryotes (Lusk, Blobel, and King 2007), which is an extension of the outer nuclear membrane, which differs in composition to the inner membrane (Dacks et al. 2016). No bacteria are known to have an ER, Golgi apparatus, or complex nuclear membrane pore complexes, and interestingly enough different sets of NCP proteins are characteristic of different groups of eukaryotes. For example, the proteins Pom121, Gp210, and the six lamin-associated proteins emerin, otefin, lamina A/C, B1, B2, Lap1 and Lap2 are unique to metazoans, whereas Pom152, Pom34, Ndc1, Nup1p and Nup2p are unique to fungi (Bapteste et al. 2005), which means that these proteins are analogous not to any prokaryotic proteins, but are analogous between different eukaryote groups. Nucleoporins show greatly divergent amino acid sequences, yet are similar secondary structures (Devos, Gräf, and Field 2014).
Compartmentalization itself is present in diverse analogous ways in several bacterial and eukaryotic groups (Devos and Reynaud 2010; Lonhienne et al. 2010); therefore they are not the products of homology. Specific to eukaryotes are P-loop NTPase motors, such as kinesins, myosins, and dyneins, which are used for directional transport in the cytoplasm (Aravind, Iyer, and Koonin 2006).
Cell division
Also, for example, some evolutionists speculate that PVC bacteria use a protein called FtsZ which plays a role in cell division. However, the phyla Chlamydia and Planctomycetes do not have this gene, and also members of the genus Prosthecobacter (from the phylum Verrucomicrobia) have both FtsZ and tubulin genes, the latter characteristic of eukaryotes (Devos and Reynaud 2010; Pilhofer et al. 2007). The proteins FtsZ and MreB show 40–50% sequence similarity across bacterial and archaeal species, and actin and tubulin are also highly conserved across eukaryotic species (75–85% sequence identity), yet these two pairs of proteins are not similar to each other sequentially, even though they are virtually identical at the level of protein folding (Erickson 2007). Coatamer proteins involved in vesicular transporters, which are also analogous between prokaryotes and eukaryotes (Dacks et al. 2016) should also be mentioned here.
Histones
Furthermore, even though some species of the phylum Chlamydia are known to contain the histone-like protein HctA and HctB, an HHPred search for homologs to the eukaryotic histone H1 failed (McInerney et al. 2011). Eukaryotic histones are also distinguished from prokaryotic ones by basic tails, composed of low-complexity sequences (Aravind, Iyer, and Koonin 2006). One would think that evolutionarily this would have been an easy add-on, and that some prokaryotes would also have them.
The translation apparatus
It is well known that the ribosome size and constituent ribosomal proteins and rRNAs differ between eukaryotes and prokaryotes. Eukaryotes have a 4200 kDA ribosome made up of a 60S and a 40S unit, whereas prokaryotes have a 2700 kDA ribosome made up of a 50S and a 30S subunit. Many post-translational elements, such as Dicer and PIWI domain nucleases are also not homologous but only analogous (Aravind, Iyer, and Koonin 2006); furthermore, the RNA interference (RNAi) machinery is also not orthologous between eukaryotes and prokaroytes (Makarova et al. 2006).
Bioenergetics and the origin of mitochondria
Prokaryotes are also limited in their cell complexity for bioenergetic reasons. A much larger cell with much more components has a lot higher energy demand to keep these components functioning. Eukaryotic cell volume has been estimated to be 15,000 times larger than that of prokaryotes on average (Lane 2011), which is also reflected in genome size. Evolutionists therefore speculate that the one major step that allowed PET is the acquisition of an energy-producing organelle.
But there are problems with these ideas as well. Even though evolutionary studies conclude that the most likely ancestor of mitochondria was α-proteobacteria, its energetic capabilities are uncertain, as compared to facultative anaerobic bacteria, such as those from the genus Rhodobacter (Esser, Martin, and Dagan 2007; Mentel and Martin 2008). Prokaryotes within prokaryotic cells indeed have been found, such as in the case of β-proteobacteria which contain γ-proteobacteria, both of which are endosymbionts of mealybugs (von Dohlen et al. 2001), but yet this still does not constitute proof for PET, but rather is evidence against it. If two different species of prokaryotes have been shown to exist with each other in endosymbiosis, the big question still remains as to what gave the impetus for transforming the gene structure of the host prokaryote into a eukaryotic one, while the genome of the endosymbiont remained prokaryotic? Since β-proteobacteria and γ-proteobacteria take part in endosymbiosis, they must have been co-existing with each other long enough for their genome structure to have undergone major changes (if we assume evolution to be true).
Second of all, within Planctomyces there are certain species which contain certain compartments which are capable of producing energy via ammonium reduction. These bacteria are called anammox bacteria, and their special compartment is called the anammoxosome, which lacks any kind of genome present in mitochondria, which it could be analogous to (van Niftrik et al. 2010). This means that endosymbiosis between prokaryotes was wholly unnecessary for bioenergy production to achieve eukaryotic cell complexity, because anammox bacteria already had their own compartment to do so, yet these bacteria never evolved into eukaryotes. Interestingly, a McInnerney et al. (2011) note, the anammoxosome has been omitted from discussion in evolutionary literature.
Discussion and Conclusion
The fact that some prokaryotes have certain eukaryotic characteristics should not be unsettling for creationists. First of all, there is nothing in the Bible which distinctly characterizes eukaryotes as the sole organisms which have nuclear membranes. The distinction between eukaryotes on the one hand having a nuclear membrane and prokaryotes on the other hand having none is a man-made construct, and therefore subject to change. Second, the presence of MC-like proteins in certain species of bacteria highlights their functional modular nature, rather than their phylogenetic relationships. Third, what we can say, is that the forest of life is made up of a more colorful spectrum of organisms, including different kinds of bacteria which happen to have analogous features to eukaryotes. These characteristics include, besides internal membranes and endocytosis, things like straight chromosomes, DNA recombination, introns, extreme polyploidy, giant size, a dynamic cytoskeleton, intercellular signaling and even endosymbionts (Huber et al. 2002; Lane 2011). Yet these characteristics have never been observed to present all together at once in any one species. Furthermore, the evolutionary literature also talks about “the apparent absence of organisms resembling putative pre-eukaryotic evolutionary intermediates” (Aravind, Iyer, and Koonin 2006). Despite the analogous similarities between prokaryotes and eukaryotes described here, there still exist a large number of minute molecular-level differences between these two cell types. Makarova et al. (2005) describe this as a most dramatic evolutionary transition, second to the emergence of life itself. Sixty of these differences have been listed in Table 2 (after Cavalier-Smith, 2009). Furthermore, according to Makarova et al. (2005) there are 4137 eukaryotic genes (almost the same number of genes in the hypothetical last eukaryotic common ancestor, LECA) which are specific to eukaryotes compared to prokaryotes. Such a large number of genes really underlines the sharp distinction between prokaryotes and eukaryotes. The majority of these genes (41 out of 54 COG clusters, 77.4%) are involved in translation, which is a process which fundamentally affects cell function. Oddly enough, some evolutionists explain the emergence of eukaryote-specific proteins via the duplication and subsequent (unobserved) “drastic acceleration” (part of a process called nonhomologous replacement) of bacterial paralogs (Aravind, Iyer, and Koonin 2006), which has never been observed. Also according to some evolutionists, these eukaryotic proteins then freeze afterwards (Makarova 2005). As to what governs these phases of sequence acceleration followed by freezing is mere speculation, and has never been observed in nature. In sum, prokaryotes with different kinds of eukaryote-like structures show the vast diversity of the kinds of living things God created. Science changes with newer and newer discoveries and observations, and creation science is up to this task.
| AAA lysine biosynthetic pathway |
| Actin and actin-related proteins (Arps 1, 2, 3) |
| Arf1 and Sar1 GTPases |
| Calmodulin, Ca++ and inositol triphosphate second messenger systems. |
| Cell cycle resetting by anaphase proteolysis |
| Cell division by actomyosin not FtsZ |
| Centrin (Ca++ contractility) |
| Centrioles and δ, ε, and η tubulins |
| Centromeres/kinetochores |
| Chromatin condensation cycle: histone phosphorylation, methylation, acetylation; heterochromatin |
| Cilia (nine doublets, dynein arms and centre pair spokes, ciliary transport) |
| Clathrin coats and adaptins |
| COPII coats |
| COPI vesicle coats |
| Delrin protein extrusion channel for ER-associated degradation (ERAD) |
| Dynein for sliding surface-attached astral microtubules and related midasin for ribosome export |
| Endosomes (early, late and multivesicular bodies) |
| Exocytosis and exocysts |
| Formins for positioning actomyosin |
| Four-module 30-subunit mediator complex regulating polII transcription |
| Golgi complex |
| Internalisation of DNA attachment sites as protoNE/roughER |
| Karyopherins |
| Lysosomes |
| Massive expansion of serine/threonine kinase controls |
| MCM replication licensing system controlled by cyclins |
| Meiosis and synaptonemal complex |
| Mitochondria |
| mRNA capping and export machinery |
| Nonsense-mediated mRNA decay |
| Nuclear envelope fusion and syngamy |
| Nuclear lamina |
| Nuclear pore complexes (NPCs) |
| Nucleolus and more complex rRNA processing (e.g. 5.8S rRNA) |
| Peroxisomes |
| Phagocytosis |
| Phosphatidylinositol/kinase signaling |
| Pinocytosis |
| Plasma membrane phosphatidylinositol anchor proteins |
| polyA transcription termination system |
| Post-transcriptional gene silencing, dicer and argonaut nucleases |
| Proteinaceous interphase nuclear matrix with bound DNA-topoisomerase II and its ability to reorganize as mitotic chromosome cores |
| Rab GTPases |
| Ran GTP/GDP cycle for directionality of NE export/import |
| Ras GTPases |
| Rho GTPases |
| Ribosome subunit nuclear export machinery |
| Separate RNA polymerases I, II and III |
| SNAREs |
| Sphingolipid synthesis |
| Spliceosomes and spliceosomal introns |
| SRβ of signal-recognition-particle receptor |
| Telomerases and telomeres |
| Trans-Golgi network |
| Tubulin: γ for centrosome and α and β for microtubules fixing it to cell surface |
| Ubiquitin and polyubiquitin labelling system |
| 3+myosins |
| 4-histone nucleosomes |
| 13 kinesins |
| 26S proteasomes with 19S regulatory subunit |
References
Aravind, L., L. M. Iyer, and E. V. Koonin. 2006. “Comparative Genomics and Structural Biology of the Molecular Innovations of Eukaryotes.” Current Opinions in Structural Biology 16 (3): 409–419.
Bapteste, E., R. L. Charlebois, D. MacLeod, and C. Brochier. 2005. “The Two Tempos of Nuclear Pore Complex Evolution: Highly Adapting Proteins in an Ancient Frozen Structure.” Genome Biology 6 (10): R85.
Bergman, J. 2001. “Does Homology Provide Evidence of Evolutionary Naturalism?” TJ 15 (1): 26–33.
Bonifacino, J. S., and J. Lippincott-Schwartz. 2003. “Coat Proteins: Shaping Membrane Transport.” Nature Reviews Molecular Cell Biology 4 (5): 409–414.
Brochier, C., and H. Philippe. 2002. “Phylogeny: A Non-Hyperthermophilic Ancestor for Bacteria.” Nature 417 (6886): 244.
Budd, A., S. Blandin, E. A. Levashina, and T. J. Gibson. 2004. “Bacterial Alpha2-Macroglobulins: Colonization Factors Acquired by Horizontal Gene Transfer from the Metazoan Genome?” Genome Biology 5 (6): R38.
Cavalier-Smith T. 2009. “Predation and Eukaryote Cell Origins: A Coevolutionary Perspective.” The International Journal of Biochemistry and Cell Biology 41 (2): 307–322.
Dacks, J. B., M. C. Field, R. Buick, L. Eme, S. Gribaldo, A. J. Roger, C. Brochier-Armanet, and D. P. Devos. 2016. “The Changing View of Eukaryogenesis—Fossils, Cells, Lineages and How They All Come Together.” Journal of Cell Science 129: 3695–3703.
de Duve, C. 2007. “The Origin of Eukaryotes: A Reappraisal.” Nature Reviews Genetics 8 (5): 395–403.
Devos, D., S. Dokudovskaya, F. Alber, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2004. “Components of Coated Vesicles and Nuclear Pore Complexes Share a Common Molecular Architecture.” PLoS Biology 2 (12): e380.
Devos, D. P., and E. G. Reynaud. 2010. “Evolution. Intermediate Steps.” Science 330 (6008): 1187–1188.
Devos, D. P., R. Gräf, and M. C. Field. 2014. “Evolution of the Nucleus.” Current Opinion in Cell Biology 28: 8–15.
Erickson, H. P. 2007. “Evolution of the Cytoskeleton.” Bioessays 29 (7): 668–677.
Esser, C., W. Martin, and T. Dagan. 2007. “The Origin of Mitochondria in Light of a Fluid Prokaryotic Chromosome Model.” Biology Letters 3 (2): 180–184.
Forterre, P., and S. Gribaldo. 2010. “Bacteria with a Eukaryotic Touch: A Glimpse of Ancient Evolution?” Proceedings of the National Academy of Sciences USA 107 (29): 12739–12740.
Forterre, P. 2011. “A New Fusion Hypothesis for the Origin of Eukarya: Better than Previous Ones, But Probably Also Wrong.” Research in Microbiology 162 (1): 77–91.
Fuchsman, C. A., and G. Rocap. 2006. “Whole-Genome Reciprocal BLAST Analysis Reveals That Planctomycetes Do Not Share an Unusually Large Number of Genes with Eukarya and Archaea.” Applied and Environmental Microbiology 72 (10): 6841–6844.
Fuerst, J. A., and E. Sagulenko. 2012. “Keys to Eukaryality: Planctomycetes and Ancestral Evolution of Cellular Complexity.” Frontiers in Microbiology 3: 167.
Fuerst, J. A., and E. Sagulenko. 2014. “Towards Understanding the Molecular Mechanism of the Endocytosis-Like Process in the Bacterium Gemmata obscuriglobus.” Biochimica et Biophysica Acta 1843 (8): 1732–1738.
Huber, H., M. J. Hohn, R. Rachel, T. Fuchs, V. C. Wimmer, and K. O. Stetter. 2002. “A New Phylum of Archaea Represented by a Nanosized Hyperthermophilic Symbiont.” Nature 417 (6884): 63–67.
Koonin, E. V. 2015. “Origin of Eukaryotes From Within Archaea, Archaeal Eukaryome and Bursts of Gene Gain: Eukaryogenesis Just Made Easier?” Philosophical Transactions of the Royal Society B Biological Sciences 370 (1678): 20140333.
Lane, N. 2011. “Energetics and Genetics Across the Prokaryote-Eukaryote Divide.” Biology Direct 6: 35.
Lee, K.-C., R. I. Webb, and J. A. Fuerst. 2009. “The Cell Cycle of the Planctomycete Gemmata obscuriglobus With Respect to Cell Compartmentalization.” BMC Cell Biology 10: 4.
Lonhienne, T. G., E. Sagulenko, R. I. Webb, K. C. Lee, J. Franke, D. P. Devos, A. Nouwens, B. J. Carroll, and J. A. Fuerst. 2010. “Endocytosis-Like Protein Uptake in the Bacterium Gemmata obscuriglobus. Proceedings of the National Academy of Sciences USA 107 (29): 12883–12888.
Lusk, C. P., G. Blobel, and M. C. King. 2007. “Highway to the Inner Nuclear Membrane: Rules for the Road.” Nature Reviews Molecular Cell Biology 8 (5): 414–420.
Makarova, K. S., Y. I. Wolf, S. L. Mekhedov, B. G. Mirkin, and E. V. Koonin. 2005. “Ancestral Paralogs and Pseudoparalogs and Their Role in the Emergence of the Eukaryotic Cell.” Nucleic Acids Research 33 (14): 4626–4638.
Makarova, K. S., N. V. Grishin, S. A. Shabalina, Y. I. Wolf, and E. V. Koonin. 2006. “A Putative RNA-Interference-Based Immune System in Prokaryotes: Computational Analysis of the Predicted Enzymatic Machinery, Functional Analogies with Eukaryotic RNAi, and Hypothetical Mechanisms of Action.” Biology Direct 1: 7.
Maynard Smith, J., and E. Szathmáry. 1995. The Major Transitions in Evolution. Oxford, United Kingdom: Freeman.
McInerney, J. O., W. F. Martin, E. V. Koonin, J. F. Allen, M. Y. Galperin, N. Lane, J. M. Archibald, and T. M. Embley. 2011. “Planctomycetes and Eukaryotes: A Case of Analogy Not Homology.” Bioessays 33 (11): 810–817.
Mentel, M., and W. Martin. 2008. “Energy Metabolism Among Eukaryotic Anaerobes in Light of Proterozoic Ocean Chemistry.” Philosophical Transactions of the Royal Society of London B Biological Sciences 363: 2717–2729.
O’Micks, J. 2015. “Cnidarians Turn Evolutionary Theory Into Jelly.” Journal of Creation 29 (3): 71–79.
Pearson, A., M. Budin, and J. J. Brocks. 2003. “Phylogenetic and Biochemical Evidence for Sterol Synthesis in the Bacterium Gemmata obscuriglobus.” Proceedings of the National Academy of Sciences USA 100 (26): 15352–15357.
Pilhofer, M., G. Rosati, W. Ludwig, K. H. Schleifer, and G. Petroni. 2007. “Coexistence of Tubulins and ftsZ in Different Prosthecobacter Species.” Molecular Biology and Evolution 24 (7): 1439–1442.
Santarella-Mellwig, R., J. Franke, A. Jaedicke, M. Gorjanacz, U. Bauer, A. Budd, I. W. Mattaj, and D. P. Devos. 2010. “The Compartmentalized Bacteria of the Planctomycetes-Verrucomicrobia-Chlamydiae Superphylum Have Membrane Coat-Like Proteins.” PLoS Biology 8 (1): e1000281.
Staley, J. T., H. Bouzek, and C. Jenkins. 2005. “Eukaryotic Signature Proteins of Prosthecobacter dejongeii and Gemmata sp. Wa-1 as Revealed By In Silico Analysis.” FEMS Microbiology Letters 243 (1): 9–14.
von Dohlen, C. D., S. Kohler, S. T. Alsop, and W. R. McManus. 2001. “Mealybug Beta-Proteobacterial Symbionts Contain Gamma-Proteobacterial Symbionts.” Nature 412 (6845): 433–436.
van Niftrik, L., M. van Helden, S. Kirchen, E. G. van Donselaar, H. R. Harhangi, R. I. Webb, J. A. Fuerst, H. J. Op den Camp, M. S. Jetten, and M. Strous. 2010. “Intracellular Localization of Membrane-Bound ATPases in the Compartmentalized Anammox Bacterium ‘Candidatus Kuenenia stuttgartiensis’.” Molecular Microbiology 77 (3): 701–715.
Wagner, M., and M. Horn. 2006. “The Planctomycetes, Verrucomicrobia, Chlamydiae and Sister Phyla Comprise a Superphylum with Biotechnological and Medical Relevance.” Current Opinions in Biotechnology 17 (3): 241–249.