
Finding Adam in the Genome: A BioLogos cover-up?
Our last post began to deliver on a promise to document and detail a very serious accusation: that BioLogos has engaged in systematic scientific error on one of their most prominent “evidences” for evolution, and that they have misrepresented the arguments for and against their claims for several years. Today’s post finishes delivering on this promise.
Wiping off the Mess?
At the end of our last post, we discussed the response of Jeff Tomkins, a young-earth creationist (YEC) with the Institute for Creation Research, to Dennis Venema’s claims about the supposed remnants of an egg-laying gene (vitellogenin; sometimes abbreviated vtg or vit or VIT) in human DNA. We observed that Venema’s claims were as illogical as the claim that the word zebra evolved from the word quota. (These two words match at 20% of their letters, just like the chicken vit1 gene and human DNA.) From February to April of 2016, Venema took Tomkins’ claims to task in a five-part series1 on the BioLogos website. In part one, Venema introduced the topic and reviewed the evolutionary evidence that he had cited in previous years. In part two, Venema continued his review, emphasizing again the relevance of shared spatial positions of genes (a subject which we’ve explored in a previous post in this series). Remarkably, rather than engage the numbers that Tomkins published (i.e., numbers that called into question whether a biologically relevant match actually exists between chicken vit1 DNA and human DNA), Venema simply doubled down on his (exaggerated and inaccurate) pictorial representation of the human-chicken vitellogenin match (Figure 1). Again, the horizontal width of either the black flecks (from the Brawand, Wahli, and Kaessmann 2008 paper2) or of the black boxes between the Chicken and Human lines (from Venema’s article) represents the amount of sequence matching between chicken and humans:
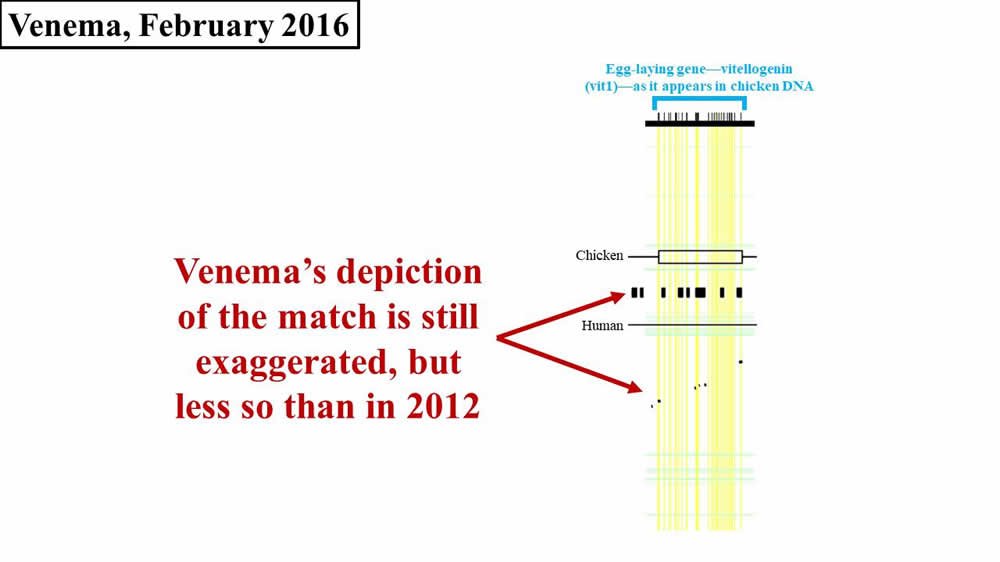
Figure 1. Comparison of 2016 Venema diagram to 2008 paper—vit1. Adapted from PLoS Biol.3 and BioLogos.4
Now compare this diagram to Venema’s depiction in 2012 (Figure 2; again, the horizontal width of either the black flecks (from the Brawand, Wahli, and Kaessmann 2008 paper5) or of the boxes on the Human line (from Venema’s article) represents the amount of sequence matching between chicken and human):
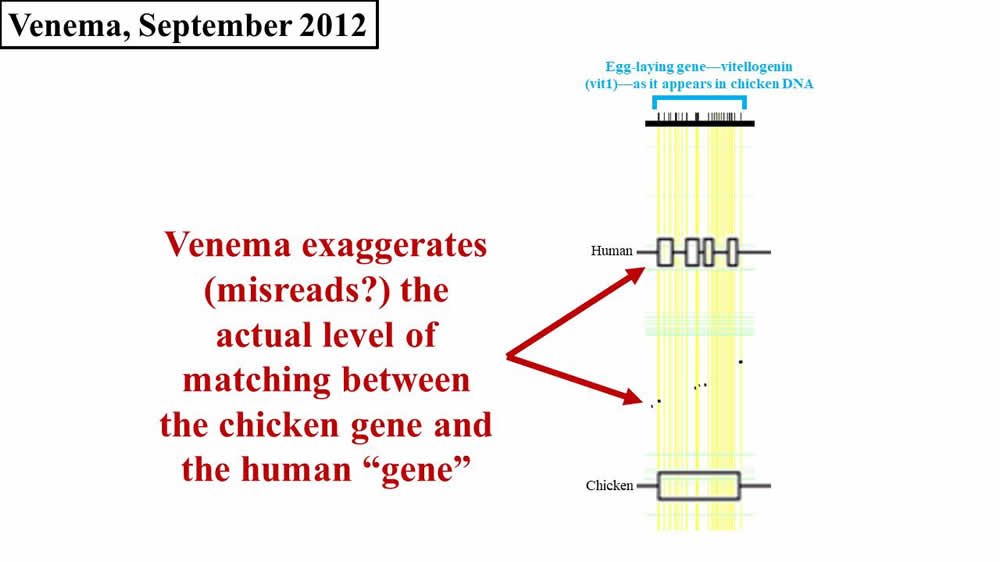
Figure 2. Comparison of later 2012 Venema diagram to 2008 paper. Adapted from PLoS Biol.6 and BioLogos.7
Do you see how Venema’s diagram changed in 2016? Why did he draw it differently in 2012? Why are his 2016 boxes different sizes? And why do his 2016 boxes still not match the original 2008 diagram?
Unlike previous years, in 2016 Venema also added his own rendition of the supposed match between the other chicken vitellogenin genes (i.e., vit2, vit3) and human DNA sequences. We discussed previously that these sequences are each about four times shorter than the vit1 sequence. Therefore, numerically, a small visual match in vit2 or vit3 is less consequential than a small match in vit1. Regardless, with respect to vit2 and vit3, Venema tried to bring the sequence matches between chicken and human before his audience (Figure 3):
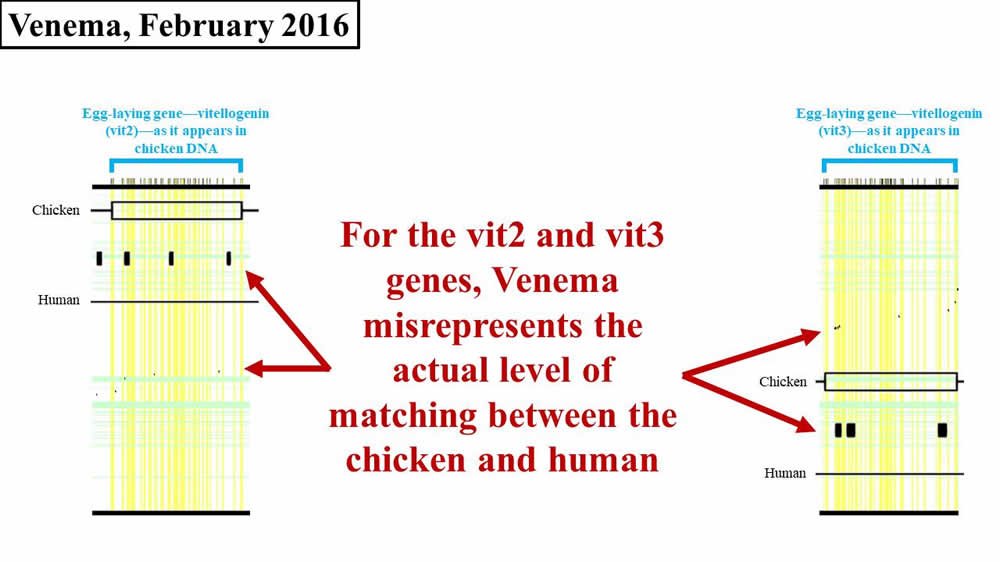
Figure 3. Comparison of 2016 Venema diagram to 2008 paper—vit2, vit3. Adapted from PLoS Biol.8 and BioLogos.9
Like his 2016 representation of vit1, Venema’s errors on vit2 and vit3 are not as egregious as they were for vit1 in 2012. Nevertheless, if you look closely at Figure 3, you’ll see that Venema’s boxes still don’t quite match the black flecks from the original paper.
In part three, Venema discusses vitellogenin in the context of other mammal species, but he provides no numbers—just additional diagrams. Venema claims that the vitellogenin gene is consistently broken in placental mammals (i.e., mammals that do not lay eggs), but not in mammals like the platypus that still lay eggs. In light of this evidence, Venema claims that evolution “has now been tested down to the molecular level—and has passed with flying colors.”10
Consistent with Venema’s failure to carefully represent and/or read the evolutionary literature on which he bases his vitellogenin claims, Venema then proceeds to misrepresent what Tomkins published. For example, Venema claims that Tomkins “focuses only on one fragment of one of the VIT pseudogenes in the human genome. This fragment is the largest continuous fragment of the human VIT1 sequence at about 150 nucleotides long. . . . Note well: this is the only sequence that Tomkins will address in his paper (!): not the other VIT1 sequence remnants surrounding this fragment.”11
Had Venema carefully read Tomkins’ paper, Venema would not have made this mistake. Tomkins clearly analyzed more than the 150-letter vitellogenin fragment:
When the human vtg pseudogene fragment [the 150 nucleotide fragment] was aligned using very liberal gapping parameters (see Materials and Methods) to the chicken genomic sequence, sequence identity was only 62%. Genomic DNA surrounding this fragment was sequentially increased three-fold in size (symmetrically) and each fragment aligned up to 36,450 bases of human genomic DNA. Sequence identity dropped as the fragment size increased, eventually leveling off to about 39% identity for a region of 36,450 bases.12
Venema missed the fact that Tomkins analyzed the 150-letter fragment—and tens of thousands of DNA letters surrounding this fragment. The 150-letter fragment is not the only sequence that Tomkins analyzed; Tomkins did indeed investigate the supposed vit1 sequence remnants surrounding this fragment.
After this egregious error in scholarship, Venema’s attacks only get worse.
Did Tomkins Miss the Main Point?
Venema also finds fault with other elements that he thinks are missing from Tomkins’ papers. Venema says that Tomkins didn’t give
even a mention of the VIT2 or VIT3 regions with their pseudogene fragments, nor the flanking DNA also found there. Similarly, the finding that these regions are shared with a wide array of other mammals is not mentioned. Tomkins has neatly bypassed the bulk of the evidence with this approach by removing the one fragment he discusses from its context, and ignoring the VIT2 / VIT3 region altogether.
With respect to vit2 and vit3, the analysis of these genes is indeed missing from Tomkins’ paper. But upon careful reflection, these omissions make sense. If Venema had carefully read the original 2008 vitellogenin paper, he would have observed that the amount of matching between humans and chickens in the vit2 and vit3 regions is even less than that for vit1. (Given Venema’s gross misrepresentation of the data in the 2008 paper, it would be no surprise if Venema missed this fact.) Not surprisingly, given the extremely low level of identity—20% (see previous post)—that Tomkins found for vit1, Tomkins didn’t even bother with vit2 or vit3.
With respect to Venema’s claim that Tomkins removed “the one fragment he discusses [i.e., the 150-letter fragment] from its context,” we’ve already documented above that this is factually false. Tomkins analyzed more than the 150-letter fragment, and Tomkins analyzed tens of thousands of DNA letters on either side of this fragment.
Pseudogenes lack experiment tests for function.
But what about the fact that Tomkins didn’t deal with vitellogenin genes in other mammals—both functional vitellogenin genes and vitellogenin “pseudogenes”? Several considerations cast this omission in a different light. First, since the match between human and chicken was so poor, why should Tomkins bother with other species? Second, with respect to functional vitellogenin genes in other species, creationists have long explained shared functional genes as consistent with common design—not just with common ancestry.13 Tomkins would have had no need to write a paper on this finding. Third, with respect to the existence of broken genes in other mammals, we might first ask if such broken genes exist. (This is what Tomkins has been asking with respect to vitellogenin “pseudogenes” in humans.) Venema again provides no numbers—just pictures. However, let’s say that bona fide pseudogenes exist in mammal species. This would simply bring the argument back to where we started—to the fact that pseudogenes lack experiment tests for function.
Thus, Tomkins “omissions” are simply logical consequences of the discoveries Tomkins’ made—and of the already published YEC literature, which Venema doesn’t seem to read.
What about Venema’s claim that Tomkins “neatly bypassed the bulk of the evidence”? Venema pulls no punches.
The true “main evidence” for the remains of VIT genes in the human genome is as we have discussed: the overall match of sequences between placental / marsupial mammals and egg-laying organisms over large spans of DNA, including flanking regions. This is the evidence that needs to be addressed—and Tomkins does not even mention it, let alone address it. It is also highly unlikely that his audience – since Tomkins is writing not for biologists but rather for laypeople who follow young-earth creationism—will be able to see this problem in Tomkins’ approach. Moreover, since Tomkins tells them that this fragment is the extent of the VIT1 pseudogene, they would have to read the original paper [the 2008 paper] by Brawand and colleagues to notice this is incorrect.14
Is the true “main evidence” the “overall match of sequences between placental/marsupial mammals and egg-laying organisms over large spans of DNA, including flanking regions” [emphasis mine]?
Let’s let Venema answer the question himself. In 2010, Venema said that “the mere presence of the mutated remains of a gene required for making egg yolk in the human genome should give even the most ardent anti-evolutionist pause.”15 By 2016, Venema’s true “main evidence” didn’t even include “the mere presence of the mutated remains of a gene required for making egg yolk in the human genome.” Why did Venema change his story? Could Tomkins’ publication in 2015 have played a role?
Perhaps Venema’s bold 2010 claims were just a temporary position that Venema later modified, subsequent to Tomkins publication. This hypothesis is testable. In 2012, Venema felt so strongly about the “the mere presence of the mutated remains of a gene required for making egg yolk in the human genome” that he posted two articles about them.16 Take a look at Venema’s diagrams. Are they depictions of “the overall match of sequences between placental / marsupial mammals and egg-laying organisms over large spans of DNA, including flanking regions”? Or are they strictly limited to the “the mutated remains of a gene required for making egg yolk in the human genome”? Do you see vit2 or vit3 show up at all in Venema’s illustrations? Or does he focus on vit1 exclusively? In fact, if the 150-letter fragment is so inconsequential in Venema’s thinking, why does he display a section of it prominently in one of his 2012 articles? Why is the “vitellogenin test”—Venema’s gauntlet for creationists—focused explicitly on “the human Vitellogenin 1 pseudogene,” and not on vit2 or vit3, or even vit1 in other mammals?
In short, from 2010 to 2012, it appears that Venema flaunted his vitellogenin argument using a very specific line of reasoning. The human-chicken vit1 match—including the 150-letter fragment—was central to Venema’s arguments. Vit2 and vit3 weren’t even present in his diagrams, let alone the vit genes in other mammal species. Then, when Tomkins exposed Venema’s arguments as scientifically deficient, Venema moved the goalposts and emphasized something different.
When Tomkins exposed Venema’s arguments as scientifically deficient, Venema moved the goalposts and emphasized something different.
Venema claims that “it is also highly unlikely that his audience—since Tomkins is writing not for biologists but rather for laypeople who follow young-earth creationism—will be able to see this problem in Tomkins’ approach.” In fact, the exact opposite is true. Tomkins published a technical paper in which he analyzed tens of thousands of DNA letters and reported a percent identity value. Venema simply drew erroneous pictures. Which author is erroneously dumbing down the data for lay audiences? We could summarize Venema’s entire approach to this question by slightly tweaking one of his own sentences: it is highly unlikely that Venema’s audience—since Venema is writing not for biologists but rather for laypeople—will be able to see this problem in Venema’s approach.
It Gets Messier
In part four of Venema’s response to Tomkins, Venema amplifies his mischaracterization of Tomkins. He takes issue with Tomkins statements about the level of DNA identity between chickens and humans in the region surrounding the supposed vit1 pseudogene. But rather than do his own analysis and report the actual level of percent identity, Venema simply reposts his erroneous picture (see above) of the 2008 findings.
To be sure, Venema recognizes that Tomkins’ is basing his numbers on original research. Venema knows that Tomkins went through effort of obtaining the raw DNA sequences and electronically comparing them himself. Consequently, Venema attacks the specific methods that Tomkins uses as “highly idiosyncratic.” Venema doesn’t give any scientific justification for this accusation. Instead, he refers his readers to previously published criticisms of Tomkins’ methodologies.
However, if you click the links that Venema supplies and then compare them to Tomkins’ published paper, you’ll find an incongruity. The published criticisms attack a method that Tomkins doesn’t even use in his analysis of vit1.17 Yet Venema concludes, “In any case, Tomkins’ claim in this instance is simply wrong, and would greatly mislead a non-specialist audience.” In fact, since Venema appears to have carefully read neither the 2008 paper nor Tomkins’ paper, the misleading is all Venema’s.
Venema also takes exception to the latter half of Tomkins’ paper. In the latter half, Tomkins supplies evidence in favor of a function for the purported human vit1 gene fragment. Venema: “The major problem with this argument is that it subscribes to a false dichotomy: that this sequence is either a VIT1 pseudogene fragment or a functional part of another gene. From an evolutionary perspective, there is no issue with it being both.”18
Venema seems to have forgotten the challenge that he laid down a couple years prior: “I would invite these [creationist] groups, all of whom . . . suggest that ‘junk DNA’ is no longer a tenable idea, to ‘take the test’ and offer an explanation for the features we observe in the human Vitellogenin 1 pseudogene.”19 Tomkins has taken Venema’s test, just as Venema requested. Why is this element of Venema’s challenge suddenly no longer important? Venema seems to have moved the goalposts again.
By part 5, Venema thinks his rebuttal to Tomkins is sufficient, and Venema moves on from the vitellogenin evidence. He says so explicitly at the end of part 4: “In the next post in this series, we’ll leave Tomkins behind and delve into the biology of how a lineage might shift from laying eggs to placental reproduction.”20
The Cracks Remain
Venema’s next published comment on vitellogenin happened in early 2017—the publication of the book, Adam and the Genome. This brings us back to where we left off our discussion of Adam and the Genome—at the end of chapter two. In chapter two, Venema continues to make the same vitellogenin arguments that he has for the last several years—with one exception. To be sure, Venema still fails to put an actual number on the percent identity between chicken vit1 and the purported human vit1 gene sequence. Instead, he still shows pictures of the supposed sequence match. But this time, Venema’s picture is different. Once again, the width of the flecks (2008 paper) or black bars (Venema’s diagram) represent the amount of DNA sequence matching between chicken and human (Figure 4):
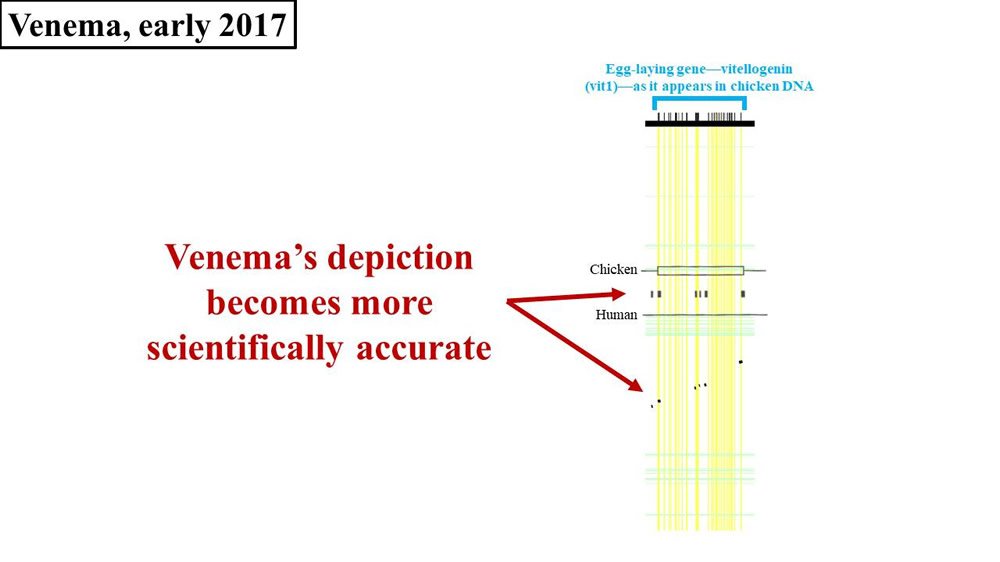
Figure 4. Comparison of 2017 Venema diagram to 2008 paper. Adapted from PLoS Biol.21 and Adam and the Genome.22
Notice that Venema’s bars appear to bear more resemblance to the flecks of sequence match shown in the original 2008 paper. Why did Venema’s diagram change? Why did it take five years for Venema to correct his science? Did Venema finally read Tomkins’ paper (instead of misrepresenting it)? If so, why is Tomkins given no mention? Why does Venema still refuse to publish any numbers on the actual percent difference between chickens and humans? If the percent identity is as low as Venema seems to now (finally) be conceding, why is he still insisting that a bona fide vit1 pseudogene exists?
Let’s consider the significance of the change in Venema’s diagrams from a different angle. Recall that this entire controversy has been a battle of numbers (Tomkins) versus pictures (Venema)—to the extent that Venema thinks his diagrams rebut Tomkins’ numbers. Not only is this unscientific, it is fatal to Venema’s position. Since Venema treats his pictures as data, Venema has essentially conceded that he doesn’t understand the scientific data / doesn’t know the scientific data. Why else would his diagrams (i.e., what he considers data) keep changing? From a scientific perspective, this is one of the strongest criticisms of Venema’s claims—and it comes from Venema himself.
Venema’s diagram is, essentially, a tacit admission of error that stretches back over most of the history of this controversy.
How does Venema’s behavior square with BioLogos’ stated commitment to “humility and gracious dialogue with those who hold other views”23 [emphasis theirs]? Will Venema humbly and publicly acknowledge his prior (public) errors? Will he graciously credit Tomkins for Tomkins’ original research? Will he correct his slanderous statements about Tomkins’ practice and character? Will he dialogue with Tomkins and learn the methods that Tomkins is actually employing? Will Venema go back and correct his earlier articles, to bring his statement on vitellogenin into agreement? Will Venema tell us what the real main evidence for the vitellogenin pseudogene is, in a manner that is consistent with Venema’s previously published statements?
Will Venema humbly and publicly acknowledge his prior (public) errors?
In the book, Venema does none of these things. Why? Because he thinks that YE creationists are liars. Consistent with this view, he closes chapter two with a selective quote from a professed YEC. This particular individual claims that evolution is well-supported by the evidence—Venema thinks that this individual is simply being “honest” about the evidence—and Venema quotes him towards this end.
What Venema doesn’t tell the reader is the rest of the story. Not surprisingly, given his professed commitment to the YEC views, the YEC individual was pressed to offer actual evidence for evolution instead of just asserting that much evidence exists. He offered none.24
With respect to vitellogenin, Venema’s public behavior is a sad reflection on the character of BioLogos. Unfortunately, given my many interactions with the members of BioLogos—in public forums; in written exchanges; in private, hours-long meals and discussions, etc.—Venema’s actions are not out of the ordinary. BioLogos has done a tremendous job publicly marketing a clean, gracious, humble image to the evangelical world; yet I’ve seen a very different (and disturbing) side in private that is consistent with Venema’s public actions. Venema’s public, documented actions should serve as a strong warning to all whose only experiences with BioLogos have been under the guise of BioLogos’ skillfully marketed (but inaccurate) message of a commitment to “humble and gracious dialogue.”
In summary, we have observed that Venema’s treatment in chapter two of the genetic evidence for evolution followed exactly what we claimed: Venema fits facts to his preconceived evolutionary conclusions. In fact, he seems to play somewhat fast and loose with the facts, as suits his purposes—making the “main evidence” for his claims one thing in one year and then another thing in another year. He also seems to have no problem misrepresenting his opponents.
We also discovered an answer to the question from a previous post (i.e., Does Venema carefully read the evolutionary literature?): it appears that Venema does not. In fact, his responses to Tomkins makes one wonder if Venema carefully reads his own claims.
Footnotes
- Dennis Venema, “Vitellogenin and Common Ancestry—Blog Series,” Letters to the Duchess blog, http://biologos.org/blogs/dennis-venema-letters-to-the-duchess/series/vitellogenin-and-common-ancestry.
- D. Brawand, W. Wahli, and H. Kaessmann, “Loss of Egg Yolk Genes in Mammals and the Origin of Lactation and Placentation,” PLoS Biol. 6, no. 3 (2008): e63, doi:10.1371/journal.pbio.0060063.
- Ibid.
- Dennis Venema, “Vitellogenin and Common Ancestry: Understanding Synteny,” BioLogos (blog), February 25, 2016, http://biologos.org/blogs/dennis-venema-letters-to-the-duchess/vitellogenin-and-common-ancestry-understanding-synteny.
- Ibid.
- Brawand, Wahli, and Kaessmann, “Loss of Egg Yolk Genes . . . .”
- Dennis Venema, “ENCODE and “Junk DNA,” Part 2: Function: What’s in a Word?,” BioLogos (blog), September 26, 2012, http://biologos.org/blogs/guest/encode-and-junk-dna-part-2/.
- Brawand, Wahli, and Kaessmann, “Loss of Egg Yolk Genes . . . .”
- Venema, “Vitellogenin and Common Ancestry: Understanding Synteny.”
- Dennis Venema, “Vitellogenin and Common Ancestry: Reading Tomkins,” Letters to the Duchess blog, March 11, 2016, http://biologos.org/blogs/dennis-venema-letters-to-the-duchess/vitellogenin-and-common-ancestry-reading-tomkins.
- Ibid.
- Jeffrey P. Tomkins, “Challenging the BioLogos Claim That a Vitellogenin (Egg-Laying) Pseudogene Exists in the Human Genome,” Answers Research Journal 8 (2015): https://answersingenesis.org/genetics/dna-similarities/challenging-biologos-claim-vitellogenin-pseudogene-exists-in-human-genome/.
- Nathaniel T. Jeanson, “Does ‘Homology’ Prove Evolution?,” Institute for Creation Research, http://www.icr.org/article/does-homology-prove-evolution.
- Dennis Venema, “Vitellogenin and Common Ancestry: Reading Tomkins.”
- Dennis Venema, “Signature in the Pseudogenes, Part 2,” BioLogos, May 17, 2010, http://biologos.org/blogs/dennis-venema-letters-to-the-duchess/signature-in-the-pseudogenes-part-2.
- Ibid. See also Dennis Venema, “Is there ‘Junk’ in Your Genome? Part 4,” BioLogos, February 17, 2012, http://biologos.org/blogs/dennis-venema-letters-to-the-duchess/understanding-evolution-is-there-junk-in-your-genome-part-4; Venema, “Encode and ‘Junk DNA,’ Part 2: Function: What’s in a Word?”
- Venema betrays another fact in these citations. Since these criticisms are out of date (for example, see the following: Jeffrey P. Tomkins, “Documented Anomaly in Recent Versions of the BLASTN Algorithm and a Complete Reanalysis of Chimpanzee and Human Genome-Wide DNA Similarity Using Nucmer and LASTZ.” Answers Research Journal 8 (2015): https://answersingenesis.org/genetics/dna-similarities/blastn-algorithm-anomaly/; Jeffrey P. Tomkins, “Analysis of 101 Chimpanzee Trace Read Data Sets: Assessment of Their Overal Similarity to Human and Possible Contamination with Human DNA,” Answers Research Journal 9 (2016): https://answersingenesis.org/genetics/dna-similarities/analysis-101-chimpanzee-trace-read-data-sets-assessment-their-overall-similarity-human-and-possible/), Venema reveals that he doesn’t pay much attention to the young-earth creation technical literature, a fact to which we alluded previously (see the following: Nathaniel T. Jeanson and Jeffrey P. Tomkins, “Why Does Mainstream Scientific Literature Ignore Conclusions from Young-Earth Creationists? Part 4,” Answers in Genesis, May 25, 2017, https://answersingenesis.org/creation-scientists/mainstream-scientific-literature-ignore-young-earth-conclusions/; Nathaniel T. Jeanson, “Creationists Are Liars? Finding Adam in the Genome with BioLogos,” Answers in Genesis, June 8, 2017, https://answersingenesis.org/creation-scientists/creationists-are-liars-finding-adam-in-the-genome-with-biologos/). If Venema doesn’t carefully do his homework on his opponents, why should we trust his statements about young-earth creation science?
- Dennis Venema, “Vitellogenin and Common Ancestry: Tomkins’ False Dichotomy,” Letters to the Duchess blog, March 24, 2016, http://biologos.org/blogs/dennis-venema-letters-to-the-duchess/vitellogenin-and-common-ancestry-tomkins-false-dichotomy.
- Dennis Venema, “Encode and Junk DNA, Part 2: Function: What’s in a Word?”
- Dennis Venema, “Vitellogenin and Common Ancestry: Tomkins’ False Dichotomy.”
- Brawand, Wahli, and Kaessmann, “Loss of Egg Yolk Genes . . . .”
- Dennis R. Venema, and Scot McKnight, Adam and the Genome: Reading Scripture after Genetic Science, Grand Rapids, MI: Brazos Press, 2017.
- “What We Believe,” BioLogos, http://biologos.org/about-us/our-mission/.
- Todd Wood, “The Evidence for Evolution,” Todd’s Blog, December 21, 2009, http://toddcwood.blogspot.com/2009/12/evidence-for-evolution.html.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- © 2024 Answers in Genesis




