Stop-or-Go DNA Codes Reveal Rampant Rule-Breaking in the Language of Life
News to Know
Abstract
When does “stop” mean “go”? Surprisingly often!
News Source
The DNA code common to all living things is not quite as uniform as previously thought. Evolutionists have seen the uniformity of the genetic code as evidence for their claim that all life evolved from a common ancestor. But there is more “babel”-like variety in microbial genomes than anyone ever suspected.
“All along, we presumed that the code or vocabulary used by organisms was universal, applying to all branches of the tree of life, with vanishingly few exceptions,” says Edward Rubin, senior author of a paper just published in Science. “We have now confirmed that this just isn't so. There is a significant portion of life that uses different vocabularies where the same word means different things in different organisms.”
Multilingual Microbes
In DNA, a genetic “word”—or codon—consists of a triplet of DNA base pairs. Most codons stand for amino acids used to build proteins. A gene is a series of codons punctuated with a “stop” codon. Until recently scientists thought all codons meant the same thing in all organisms. But now a study has shown that the “periods” at the ends of “genetic sentences” are routinely re-purposed in many microbial genomes. Codons ordinarily read as “stop; this gene ends here” do not, it turns out, always mean “stop.”
Some microbes—particularly friendly bacteria that inhabit the human body—see things differently. For instance, some read one of the “stop” codons as “insert glycine here.” A codon with an alternative meaning also appears in at least one microbial eukaryotic ciliate. And bacteriophages—viruses that insert their genomes into bacterial genomes in order to replicate themselves—can even re-purpose a couple of these stop signals to code for an amino acid that suits their needs.
Microbial Dark Matter
Microbes accustomed to cozy positions in our bodies or to extreme conditions in hot undersea vents resist being tamed and tested in petri dishes. This DNA innovation was discovered in a U.S. Department of Energy Joint Genome Institute project aimed at uncovering the secrets of this “microbial dark matter”—the enormous chunks of the microbial world that defy conventional efforts to culture them in the laboratory. We think we know a lot about microbes, but 99% of microbial species stubbornly conceal their secrets outside the veil of the lab door.
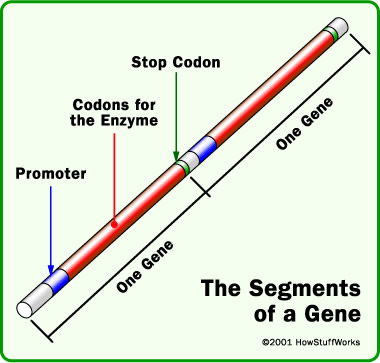
A gene contains of a series of codons, each prescribing an amino acid to be incorporated into a protein. Each gene is punctuated at the end by a “stop” codon. Image from HowStuffWorks.3
So how can you study a microbe that won’t sit still and grow while you watch? You have to directly study the genes in individual cells and in environmental samples (aka metagenomics), a technologically more difficult prospect. But metagenomics has revealed previously unsuspected microbial diversity.
“The tools of metagenomics and single-cell genomics, with which we determine the genetic blueprints of microbes without the need to grow them in the laboratory, provide us a window into the unexplored, uncultured microbial world,” Rubin explains. “The metaphor we use is that up until very recently we have just been looking under the lamppost for new life, studying organisms that we can grow in the laboratory while we know most microbial life is very resistant to being grown in the lab. In this project, using metagenomics and single-cell genomics to explore uncultured microbes, we really had the opportunity to see how the genetic code operates in the wild. It is helping us get an unbiased view of how nature operates and how microbes manage our planet.”
Non-Canonical Codons
Stop signals indicate where one gene ends and the next begins. Only three of the 64 DNA codons—nicknamed Amber, Opal, and Ochre—are “stop” codons. Natalia Ivanova, lead author of the report in Science, discovered some bacteria with extraordinarily short genes—only 200 base pairs long compared to the usual 800-900. But they don’t really have pint-sized genes; instead, these bacteria simply march to a different drummer. In their language, “Opal” doesn’t mean “stop”; it means “glycine,” one of the amino acids.
“When trying to interpret the sequence of these bacteria using the canonical codon table, Opal, normally interpreted as a stop sign, resulted in the bacteria having unbelievably short genes,” Rubin explains. “When Natalia applied a different vocabulary where Opal, instead of being interpreted as a stop, was assumed to encode the amino acid glycine, the genes in the bacteria suddenly appeared to be of normal length. Their interpretation of the finding was that ‘Opal-recoded’ organisms, instead of stopping, incorporated an amino acid into the polypeptide, which kept growing and eventually produced normal-sized proteins.”
Rare or Routine?
Was this the Rosetta stone of molecular genetics, or just a fluke? How common is this variable DNA reading? The researchers found examples in over 1,700 genomes from marine, freshwater, and terrestrial ecosystems worldwide as well as the human mouth and gut. Rubin says, “We were surprised to find that an unprecedented number of bacteria in the wild possess these codon reassignments, from ‘stop’ to amino-acid encoding ‘sense,’ up to 10 percent of the time in some environments.”
Alternative readings were found for all three “stop” codons. Many bacteria read “Amber” as an amino acid. One eukaryotic ciliate defines “Ochre” as an amino acid. Bacteriophages re-purpose “Amber” and “Opal.”1 Some of the co-opted codons are even used to encode amino acids not ordinarily included in proteins, selenocysteine and pyrrolysine. Yale University biochemist Dieter Söll comments, “My greatest excitement about this paper is really that we now know that there is a great frequency of stop codon changes. I would imagine that if the biochemistry would be done on all these examples [of organisms that have reassigned stop codons], one would very likely find additional genetically encoded non-standard amino acids.”2
Phages Follow Their Own Rules
Bacteriophages don’t follow the rules microbiologists thought they did. Codon re-purposing is apparently part of the toolkit these viruses use in taking over bacterial hosts. Bacteriophages suppress the host’s protective mechanisms and change the meaning of a codon from “stop” to an amino acid suiting its needs.
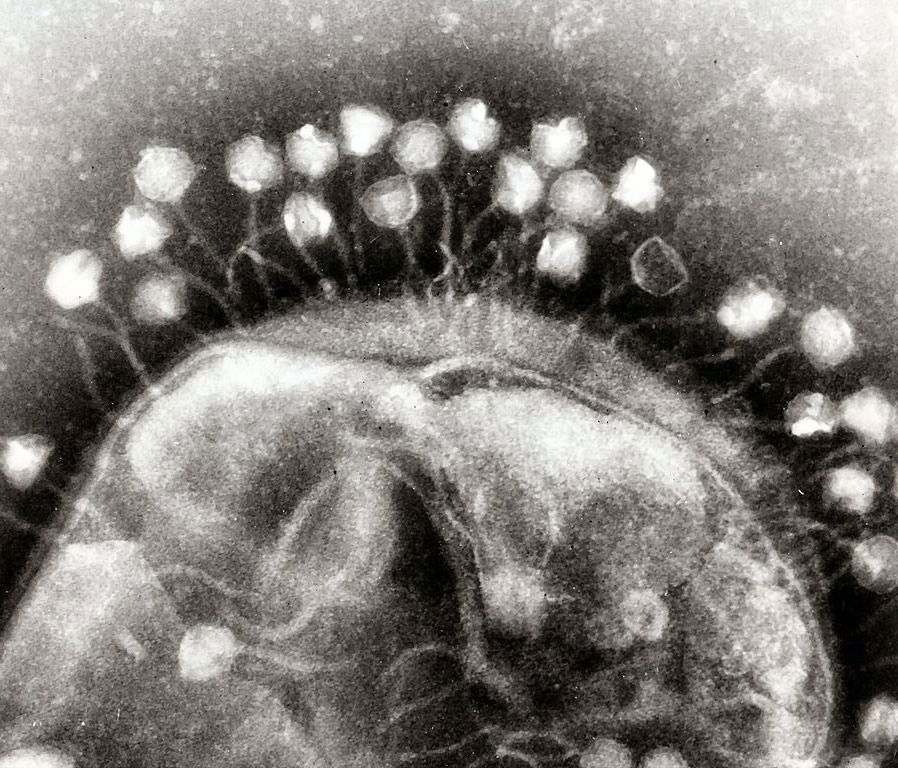
These bacteriophages are attacking a bacterium, co-opting its genetic replicating machinery to manufacture more phages. New research reveals that some viral phages can change the meaning of genetic “stop signs” in the bacterial DNA. Image by Webridge, via Wikimedia Commons.
“The established dogma was that phage needed to employ the exact genetic code that the host cell uses, otherwise, whatever DNA they inject wouldn't be properly translated,” Rubin explains. “But we observed phage with codon vocabularies that did not match any we found in their bacterial hosts. We scratched our heads at this result, because we wondered about what was up with the host. The dogma tells us that the phage to need [sic] to share the same code as the host, but we saw no Amber in bacteria. So what were these phage doing?”
“Phage apparently don't really ‘care’ about the codon usage of the host. They have ways to get around that, and in fact they use differences to attack the host,” Rubin explains. “We call this strategy ‘codon warfare.’” Biologists who genetically engineer organisms with a “firewall” consisting of “stop” codons will in future need to be aware that their genomic security could also be breached by this mechanism.
My Father’s World
God designed a perfectly good (Genesis 1:31) world in the beginning. All organisms God originally createdwould have therefore served valuable roles. While 6,000 years of sin’s curse has perverted much of that good world, the abundance of microbes and viruses in all ecosystems, even inside our bodies, as well as the valuable roles they play are consistent with a biblical understanding of our origins.
While evolutionary thinking would expect to find primitive simplicity in microbes, these DNA code variations represents a level of complexity that defies evolutionary expectations. Commenting on the study, Institute for Creation Research geneticist Dr. Jeffrey Tomkins, who recently reported on how the complexity and diversity of molecular motors violates evolutionary expectations, says, “It only complicates the picture for the evolutionist and makes the schema of microbial life more complex. Single celled organisms are once again defying the simple-to-complex inferred evolutionary pattern of life.”4
In human language, word meaning is often determined by context. Why should biological language—the genetic language of life created by our infinitely wise Creator—not use variant readings to store and even adjust the information in microbes, many of which play vital roles in the environment and in our bodies. Yale University bioengineer Farren Isaacs points out, “This study highlights the malleability of the genetic code.”
What does this discovery mean for human health and welfare? Although only a tenth of the more-than-31,000 samples surveyed came from inside the human body, over half of the polyglot microbes Rubin’s team discovered were part of the human microbiome. Our microbiome includes microbes that routinely inhabit our bodies in a mutually symbiotic relationship. They help keep us healthy, reminding us of our Creator’s care. We may yet discover that codon re-purposing is a mechanism God put in place to equip microbes to adapt and adjust to their host environments in order to best serve the roles He designed for them.
Further Reading
- “Partners in Slime”: Mucus and Our Viral Friends
- New Letters for Life’s DNA Alphabet Build on God’s Design
- The Matrix—Life's Support System
- The Organosubstrate of Life
- Microbes and the Days of Creation
- Cells’ Molecular Motor Diversity Confounds Evolution
- Duons: Parallel Gene Code Defies Evolution
- Dual-Gene Code Discovery Highlights Designed Biocomplexity
For More Information: Get Answers
Remember, if you see a news story that might merit some attention, let us know about it! (Note: if the story originates from the Associated Press, FOX News, MSNBC, the New York Times, or another major national media outlet, we will most likely have already heard about it.) And thanks to all of our readers who have submitted great news tips to us. If you didn’t catch all the latest News to Know, why not take a look to see what you’ve missed?
(Please note that links will take you directly to the source. Answers in Genesis is not responsible for content on the websites to which we refer. For more information, please see our Privacy Policy.)
Footnotes
- Natalia N. Ivanova et al., “Stop Codon Reassignments in the Wild,” Science 344, no. 6186 (May 23, 2014): 909–913, doi:10.1126/science.1250691.
- Ruth Williams, “When Stop Means Go,” TheScientist, May 22, 2014, http://www.the-scientist.com ?articles.view/articleNo/40048/title/When-Stop-Means-Go.
- Marshall Brain, “How Cells Work,” HowStuffWorks, http://science.howstuffworks.com/life/cellular-microscopic/cell7.htm.
- Personal communication (e-mail).
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- © 2024 Answers in Genesis