
Chapter 24
Beneficial Mutations
Epigenetics, an exciting field of science, displays the intelligence and providence of God to help organisms adapt and survive in a fallen world which helps explain beneficial mutations.
Many claim that beneficial mutations provide examples of “evolution in action.” These mutations supposedly result in the formation of “major innovations” and “rare and complex traits”1 that over time have resulted in the evolution of all living things from a common ancestor. However, analyses of these mutations show they only result in variations in pre-existing traits, traits that organisms already possess, and cannot result in the origin of novel traits necessary for molecules-to-man evolution.
Novelty Required for Beneficial Mutations
For a simple, single-celled ancestor to evolve into a human over billions of years, novel traits must be gained.
For a simple, single-celled ancestor to evolve into a human over billions of years, novel traits must be gained. New anatomical structures — like brains, arms, and legs — and new functions — like cardiovascular and muscle activities — must be acquired. Regardless of whether this is proposed to occur through beneficial mutations that result in the addition of new DNA, changes in existing DNA, or through other mechanisms, there must be a way to add novel traits. However, all observed mechanisms, including beneficial mutations, do just the opposite — they cause the loss of or slight variation in pre-existing traits.2 Beneficial mutations and other mechanisms cannot account for the origin of novel traits of the type necessary for molecules-to-man evolution. In a paper entitled “A Golden Age for Evolutionary Genetics? Genomic Studies of Adaptation in Natural Populations,” the authors (who are evolutionists) agree that the lack of mechanisms to add novel traits is a problem: “Most studies of recent evolution involve the loss of traits, and we still understand little of the genetic changes needed in the origin of novel traits.”3
In this paper, the scientists give many examples of variations in organisms such as pattern changes in butterfly wings, loss of bony structures in stickleback fish, loss of eyes in cavefish, and adaptations to temperature and altitude. But none of these examples involve the origin of novel traits necessary to evolve into a different kind of organism. Again, they realize this problem and state, “. . . over the broad sweep of evolutionary time what we would really like to explain is the gain of complexity and the origins of novel adaptations.”4
Their frustration with the lack of evidence for “novelty-gaining” mechanisms like beneficial mutations sinks to apparent desperation when they state, “Of course, to some extent the difference between loss and gain could be a question of semantics, so for example the loss of trichomes [hair-like appendages on flies] could be called gain of naked cuticle.”5 The authors have decided that the whole loss/gain issue is merely one of semantics! In order to get the gain required by molecules-to-man evolution they will just change the wording and say it is a “gain of loss.”
That’s equivalent to a person who has suddenly lost all their money saying, “I’ve not lost money; I’ve just gained poverty!” While it makes the person sound optimistic, it doesn’t change the fact that they have lost all their money. In the same way, an organism doesn’t gain novel traits needed to evolve into something else — instead, organisms lose traits or develop variations in pre-existing traits. It doesn’t matter how evolutionists choose to say it; there is still no mechanism that results in the origin of novel traits required for molecules-to-man evolution.
Do Beneficial Mutations Exist?
While beneficial mutations may not result in the origin of novel traits necessary to go from molecules to man, they do exist . . . sort of. Let me explain. It is more appropriate to say that some mutations have beneficial outcomes in certain environments. Mutations are context dependent, meaning their environment determines whether the outcome of the mutation is beneficial. One well-known example of a proposed beneficial mutation is antibiotic resistance in bacteria.6 In an environment where antibiotics are present, mutations in the bacterial DNA allow the bacteria to survive. However, these same mutations come at the cost of damaging the normal functions of the bacteria (such as the ability to break down nutrients). If the antibiotics are removed, the antibiotic resistant bacteria typically do not fare as well as the normal (or wild-type) bacteria that have not been affected by mutations. Thus, the benefit of any given mutation is not an independent quality, but rather a dependent quality based on the environment.
There is no question that mutations can be beneficial in certain environments, but do they lead to the origin of novel traits of the type necessary for molecules-to-man evolution?
Another common example of a supposed beneficial mutation, this time in humans, is individuals that are resistant to infection with HIV. These people have a mutation that prevents HIV from entering the white blood cells and replicating, making them unlikely to develop AIDS. However, studies have shown these individuals may be at a higher risk of developing illness associated with West Nile virus7 and hepatitis C8 (also caused by a virus). Again, we see that the mutations are only beneficial in a given environment, such as if the person were exposed to HIV. It is possible that the mutations would not be beneficial in other environments such as if the person were exposed to West Nile virus. The benefit of any given mutation is a dependent quality based on the environment.
There is no question that mutations can be beneficial in certain environments, but do they lead to the origin of novel traits of the type necessary for molecules-to-man evolution? Let’s look at several examples commonly used to support this idea and the problems with them.
Proposed Beneficial Mutations in Bacteria
Richard Lenski and the Citrate Mutation in E. coli
In 1988, Dr. Richard Lenski, an evolutionary biologist at Michigan State University, began culturing 12 identical lines of Escherichia coli (a common gut bacteria). Over 50,000 generations and 25 years later, the experiment continues. Lenski has observed many changes in the E. coli as they adapt to the culture conditions in his lab. For example, some lines have lost the ability to break down ribose (a sugar),9 some have lost the ability to repair DNA,10 and some have reduced ability to form flagella (needed for movement).11 In other words, they’ve gotten lazy as they’ve adapted to life in the lab! If they were grown in a natural setting with their wild-type (normal) counterparts, they would not stand a chance in competing for resources.
In 2008, Lenski’s lab discovered another change in one of their lines of E. coli. A New Scientist writer proclaimed, “A major innovation has unfurled right in front of researchers’ eyes. It’s the first time evolution has been caught in the act of making such a rare and complex new trait.”12 But was this change really the formation of “a rare and complex new trait”?
Normal E. coli has the ability to utilize citrate as a carbon and energy source when oxygen levels are low. They transport citrate into the cell and break it down. Lenski’s lab discovered that one of their E. coli lines could now utilize citrate under normal oxygen levels.13 It’s easy to see that this was not “a major innovation” or the “making of a rare and complex new trait” because the normal E. coli already has the ability to transport citrate into the cell and use it! This was simply a beneficial outcome of mutations that changed under what conditions citrate was used by E. coli.14 The mutations caused the alteration of a pre-existing system, not the origin of a novel one. There is a lot of citrate in the medium that the bacteria are grown in, and since other carbon sources are not plentiful, the bacteria have merely adapted to the lab conditions.
Lenski stated, “It is clearly very difficult for E. coli to evolve this function. In fact, the mutation rate of the ancestral strain . . . is immeasurably low. . . .”15 If developing the ability to utilize citrate under different conditions by altering the pre-existing citrate system is so rare, then how much more improbable is it to believe that similar beneficial mutations can lead to the origin of novel traits necessary for dinosaurs to evolve into birds!
Nylon-Digesting Mutation in Bacteria
In the mid-1970s, bacteria (Arthrobacter sp. K172) were discovered in ponds with wastewater from a nylon factory that could digest the byproducts of nylon manufacture. Nylon is a synthetic polymer that was first produced in the 1940s, thus, the ability of bacteria to break down nylon must have been gained in the last few decades. Many evolutionists touted that the bacteria’s ability to break down nylon occurred through the gain of new genes and proteins. In a 1985 article entitled “New Proteins Without God’s Help,” the author explained testing that supposedly showed the bacteria’s ability to break down nylon was due to the formation of new proteins, not the modification of pre-existing ones.16 In conclusion he stated, “All of this demonstrates that . . . the creationists . . . and others who should know better are dead wrong about the near-zero probability of new enzyme formation. Biologically useful macromolecules are not so information-rich that they could not form spontaneously without God’s help.”17
Does this mean that biblical creationists should run screaming and stick our heads into the sand?
Does this mean that biblical creationists should run screaming and stick our heads into the sand? No. In 2007, genetic analyses of Arthrobacter sp. K172 showed that no new genes or proteins had been added that resulted in the ability of the bacteria to break down nylon.18 Instead it was discovered that mutations in a pre-existing gene resulted in a protein that is capable of breaking down nylon. The protein, known as EII, normally breaks down a substance very similar to nylon. Slight alterations in what is called the “active site” of the protein (where the activity of breaking down the substance occurs) changed its specificity such that it could now also break down nylon. No changes occurred of the type necessary to go from molecules to man, just a “tweak” in a gene and protein whose normal function is to break down something very similar to nylon. Again, we see the alteration of a pre-existing gene and protein, not the origin of new ones. Information-rich molecules like DNA and protein cannot spontaneously form — they do need “God’s help.”
Barry Hall and the ebg Mutation in E. coli
Beginning in the 1970s and continuing into the 1990s, Dr. Barry Hall, professor emeritus of the University of Rochester, New York, did extensive work in the field of what has been termed adaptive or directed mutations. According to evolutionary ideas, mutations are random changes in the DNA that may or may not be beneficial to an organism in its environment. However, research from scientists like Hall has indicated that adverse environmental conditions, like starvation, may initiate mechanisms in bacteria that result in mutations that specifically allow the bacteria to survive and grow in a given environment. These changes do not appear to be random in respect to the environment, thus the term directed or adaptive mutations.
There are two reasons why adaptive mutations are problematic for evolution. First, the mechanisms in bacteria for generating adaptive mutations are specifically responding to the environment. The changes are goal-oriented, allowing the organism to adapt and survive by alteration of pre-existing traits. A second reason is that the mechanisms resulting in adaptive mutations (which appear to be a very common type of mutation in bacteria) set limits on the genetic change possible and cannot account for the origin of novel traits.
E. coli can break down the sugar lactose to use as a food source. Hall was able to mutate a strain of E. coli such that it lost the ability to break down lactose.19 He then put the mutant E. coli in a starvation situation where lactose was the only food source. In order to survive, the E. coli either had to develop the ability to break down lactose or die. After a period of time, E. coli developed the ability to break down lactose. How did E. coli do this? Were new genes and proteins added to allow this to happen?
No. Genetic analyses showed that mutations had occurred in a group of pre-existing genes named ebg. These genes are in normal E. coli and produce proteins that very weakly break down lactose. The genes were also present in Hall’s mutant E. coli (he mutated only the primary set of genes used for lactose breakdown not the ebg genes). In response to the starvation conditions, mechanisms were initiated in the bacteria that resulted in mutations in the ebg genes that produced proteins with enhanced ability to break down lactose well enough that the mutant bacteria could survive. No new or novel traits were gained, there was merely the alteration of a pre-existing trait that allowed the bacteria to adapt and survive.
Interestingly, Hall theorized that if both the primary set of genes needed for lactose breakdown and the ebg genes were made non-functional (through mutations) that adaptive mutations would occur in other genes resulting in E. coli once again developing the ability to break down lactose.20 However, “despite extensive efforts,” Hall was unable to get E. coli that could survive on lactose. They did not survive because adaptive mutations only make limited changes. Ebg genes in E. coli already possess the ability to break down lactose, adaptive mutations enhanced this ability. Adaptive mutations cannot make possible the origin of lactose breakdown from genes whose functions are not as similar.
Despite the evidence, Hall concluded this aspect of his research by saying, “Obviously, given a sufficient number of substitutions, additions, and deletions, the sequence of any gene can evolve into the sequence of any other gene.”21 But Hall’s own experiments showed otherwise — a gene cannot just become a completely different gene; adaptive mutations are limited. Mutations can cause changes in pre-existing traits, but observable mechanisms, such as adaptive mutation, cannot account for the origin of novel traits necessary for molecules-to-man evolution.
Proposed Beneficial Mutations in Animals
TRIM5-CypA Mutation in Monkeys
The TRIM5 gene is found in humans, monkeys, and other mammals. The protein produced from this gene binds to the outer covering (capsid) of retroviruses (like HIV) and prevents them from replicating inside cells, thus essentially preventing the spread of infection. A portion of the TRIM5 gene (C-terminal domain) seems especially variable and may confer resistance to different types of viruses.22 In 2004, it was discovered that owl monkeys (Aotus sp.) have a unique version of the TRIM5 gene that appears to be a fusion of this gene to the nearby CypA gene.23 The CypA gene can produce a protein that also binds to the outer covering of viruses, including HIV. Thus, the TRIM5-CypA fusion protein has the antiviral activity of TRIM5 coupled to the HIV recognition of CypA and the fused protein was able to prevent infection from HIV. (A similar fusion gene/protein has also been discovered in certain species of macaques.)24
New Scientist writer Michael Le Page in an article entitled “Evolution Myths: Mutations Can only Destroy Information,” stated in regard to this mutation, “Here, a single mutation has resulted in a new protein with a new and potentially vital function. New protein, new function, new information.”25 But is this really a new protein with a new function?
No. TRIM5-CypA is the fusion of two pre-existing genes producing a fused protein. The fusion doesn’t change the function of TRIM5 or CypA, so there is no new function. The addition of CypA merely allows TRIM5 to recognize a different group of viruses and exert its antiviral activity against those viruses. This fusion does not result in the origin of a novel trait of the type necessary for molecules-to-man evolution.
Gene Duplication, Mutation, and “New” Genes and Functions
Evolutionists often cite gene duplication, followed by subsequent mutation of the duplicated gene, as a mechanism for adding new genes with new functions to organisms. The idea is that the duplicated gene is free to mutate and gain new functions because the original copy of the gene can still perform the original function. Evolutionary biologist Dr. Sean Carroll, referring to his work on gene duplication in yeast, stated, “This is how new capabilities arise and new functions evolve. This is what goes on in butterflies and elephants and humans. It is evolution in action.”26 However, a deeper look at a couple of examples of gene duplication and mutation show exactly the opposite — the complete impotence of these mechanisms to explain the origin of novel traits necessary for molecules-to-man evolution.
RNASE1 and 1B in Monkeys
The diet of most monkeys consists of fruit and insects; however, the colobine monkeys predominantly eat leaves. These monkeys have a special foregut that harbors symbiotic bacteria that help in the digestion of the leaves. RNASE1 is a digestive enzyme in colobines that breaks down RNA from the bacteria in the foregut. This results in the efficient recycling of phosphorus and nitrogen that are used in the production of the monkey’s own proteins and nucleic acids like DNA and RNA.
It has been shown that some colobines have two RNASE genes—RNASE1 and RNASE1B.27 RNASE1B is proposed to be a duplication of the gene RNASE1. There are several differences in the genes and the proteins produced, however, the function remains the same. Both enzymes break down RNA, but the changes in RNASE1B allow it to break down RNA in more acidic conditions such as those found in the foregut of the monkeys. The authors of one study of the RNASE1 genes commented, “Gene duplication has long been thought by evolutionary biologists to be the source of novel gene function. . . . We believe our data to be another example that do not support this hypothesis.”28 Other authors of similar research indicate: “Taken together, our results provide evidence of the important contribution of gene duplication to adaptation of organisms to their environments.”29 The differences (caused by mutations) in the RNASE1B gene appear to enhance the pre-existing function of the original RNASE1 gene, resulting in adaptation, and do not represent the type of mutation necessary for the origin of novel traits needed for molecules-to-man evolution.
Antifreeze Proteins in Fish
Antifreeze proteins (AFPs) prevent the growth of ice crystals in organisms that live in very cold environments such as the Arctic and Antarctica. There are five classes of these proteins found in fish. AFP type III is found in the Antarctic zoarcid fish. The AFPIII gene is proposed to be a duplication of a portion of the SAS (sialic acid synthase) gene.30 The SAS gene is responsible for the synthesis of sialic acids (found on cell surfaces) but also has an antifreeze function. Mutations in the AFPIII gene (a duplicate copy of a portion of the SAS gene) appear to have further enhanced the antifreeze function.
One of the authors of the study on the formation of the AFPIII gene commented, “This is the first clear demonstration . . . [of ] the underlying process of gene duplication and the creation of a completely new function in one of the daughter [duplicate] copies.”31 But the AFPIII protein does not have a “completely new function”! Instead the AFPIII gene is likely the result of a duplication of a portion of the pre-existing gene with mutations that enhanced the SAS gene’s pre-existing antifreeze function. Once again, we see that the differences (caused by mutations) in the AFPIII gene appear to enhance the preexisting antifreeze function of the original SAS gene resulting in adaptation to the environment and do not represent the type of mutation necessary for the origin of novel traits needed for molecules-to-man evolution.
Beneficial Mutations from a Biblical Creation Perspective
In every example, it appears that the mutations help organisms in adapting to a specific environment.
The previous examples show that there can be beneficial outcomes to mutations. However, these mutations can only alter pre-existing traits; they cannot result in the origin of novel traits necessary for molecules-to-man evolution. In every example, it appears that the mutations help organisms in adapting to a specific environment. This is easily seen in bacteria when they are faced with limited food choices and must gain the ability to break down a different nutrient or die. It is also seen in animals like monkeys and fish that have essentially become more specialized for eating a particular diet or living in a particular environment.
But are these mutations random in respect to the environment? On the Evolution 101 website, sponsored by the University of California Museum of Paleontology, it states:
The mechanisms of evolution — like natural selection and genetic drift — work with the random variation generated by mutation. (emphasis in original)
For example, exposure to harmful chemicals may increase the mutation rate, but will not cause more mutations that make the organism resistant to those chemicals. In this respect, mutations are random — whether a particular mutation happens or not is generally unrelated to how useful that mutation would be.32
The basis of molecules-to-man evolution is random mutation in conjunction with other mechanisms like natural selection. However, mutations with beneficial outcomes do not appear to be random or at least the mechanisms generating the mutations are not random. From a biblical creation perspective, this could be a type of adaptive variation that God has designed in organisms to allow them to survive in a world dramatically changed by the Fall and Flood. Rather than the changes being random, organisms have been “pre-programmed” to change in response to their environment.
These types of adaptive traits may be the result of what creationists have termed mediated design. Several creation scientists describe it this way:
God specifically designed the created kinds with genes [in the DNA] that could be turned on to help them adapt to new environments. In other words, the Creator continues to accomplish His purpose for organisms after creation, not by creating something new, but by working through existing parts that were designed during Creation Week. An analogy is the manufacturer of a fully equipped Swiss army knife, who stores within the knife every tool a camper might need as he faces the unknown challenges of wilderness living.33
God designed adaptive traits to be expressed only under certain conditions to allow microbes, animals, plants, and humans to fill the earth as environments changed over time (Genesis 1 and 8:16–19). Thus, God programmed organisms with mechanisms that would be triggered under certain conditions that would then modify pre-existing traits to allow organisms to survive and thrive in new environments. Possible mechanisms to accomplish this are seen in the previous examples with directed mutations (ebg and E. coli) and duplication followed by mutation (RNASE1 and 1B in monkeys). Another exciting area of modern genetics research is the role of epigenetics in modifying how genes and, thus, the physical traits, are expressed. Epigenetic markers, chemical tags on DNA, have been shown to be heritable and may be a way to pass on modified traits to future generations (see postscript). Understanding the God-given ability of organisms to change and adapt is an active area of creation research.
What adaptive variations can’t do is change one kind of organism into a completely different kind of organism because they do not result in the origin of novel traits needed for this type of change.
But what adaptive variations can’t do is change one kind of organism into a completely different kind of organism because they do not result in the origin of novel traits needed for this type of change. This is consistent with Scripture because God created animals and plants according to their kind (usually at the family level in modern classification schemes).34 The inference from Scripture is that animals were to reproduce according to their kind (Genesis 1, 6, and 8). We observe mechanisms that allow animals and plants to adapt but not evolve into different kinds of organisms.
So why, in spite of all the evidence to the contrary, do many scientists, who are unbelievers, argue that beneficial mutations are a valid mechanism (as evidenced by their quotes) to account for the origin of novel traits resulting in molecules-to-man evolution? Paul says that God can be known through what He has created (Romans 1:20), but just before that Paul states why people don’t acknowledge God as the Creator: “For the wrath of God is revealed from heaven against all ungodliness and unrighteousness of men, who suppress the truth in unrighteousness” (Romans 1:18, emphasis added). Just as Pharaoh hardened his heart repeatedly (1 Samuel 6:6), today, people’s hearts have been hardened in their willful rebellion against God. They want to continue in their sin and will go to extremes to “deny the obvious” and reject God as the Creator.
God, in His mercy, compassion, and grace, designed living organisms with the ability to adapt and fill, survive and thrive in a fallen world. We look forward to the day when all life will be restored to perfection and the wolf will live with the lamb, the lion will eat straw like an ox, and a baby will play by the cobra’s hole (Isaiah 11:6–8).
Postscript: Beneficial Mutations & Epigenetics
All our lives, we’ve heard that our physical makeup is determined by our genes, not environment. But the science of epigenetics is forcing scientists to rethink their assumptions.
You’re probably familiar with the phrase, “You are what you eat.” But did you know that you are also what your mother and grandmother ate? The budding science of epigenetics shows that our physical makeup is about much more than inheriting our mother’s eyes or our father’s smile.
We are accustomed to thinking that the only thing we inherit from our parents are genes — packets of information in DNA that give instructions for proteins. These genes determine our physical traits such as hair and eye color, height, and even susceptibility to disease.
But we also inherit specific “modifications” of our DNA in the form of chemical tags. These influence how the genes express our physical traits. The chemical tags are referred to as “epigenetic” markers because they exist outside of (epi-) the actual sequence of DNA (-genetics).
Let me use an analogy to explain. The following sentence can have two very different meanings, depending on the punctuation used. “A woman, without her man, is nothing” or “A woman: Without her, man is nothing.” Perhaps it’s a silly illustration, but it gets the point across.
The words of both sentences are the same, but the meaning is different because of the punctuation. The same is true for DNA and its chemical tags. The sequence of DNA can be identical but produce different results based on the presence or absence of epigenetic markers. For example, identical twins have the same DNA sequence but can have different chemical tags leading one to be susceptible to certain diseases but not the other.
Parents can pass down epigenetic markers for many generations or their effect can be short-lived, lasting only to the next generation.
Parents can pass down epigenetic markers for many generations or their effect can be short-lived, lasting only to the next generation. Either way, the changes are temporary because they do not alter the sequence of DNA, just the way DNA is expressed.
What does this mean in practice? Your behavior, including the food you eat, could change how your body expresses its DNA. Then those changes — for good or bad — could be passed to your children! If you do something to increase your susceptibility to obesity or cancer or diabetes, your children could inherit that from you.
In one experiment, mice from the same family, which were obese because of their genetic makeup, were fed two different diets. One diet consisted of regular food. The other diet consisted of the same food but contained supplements that were known to alter the chemical tags on DNA.
Normally when these mice eat regular food they produce fat offspring. However, the mice that ate the same food with the supplements produced offspring that were normal weight. The parents’ diet affected their offspring’s weight!
Scientists are still trying to understand the details. The epigenetic markers that were modified by the food supplements appear to have “silenced” genes that encourage appetite. The parents’ environment — in this case, the food they ate before becoming parents — affected the weight of their offspring.
Certain types of medicine have also been suspected of causing changes in epigenetic markers, leading to cancer in the offspring of women who took the medicine. For example, a type of synthetic estrogen prescribed to prevent miscarriages has been linked to an increased number of cancers in their daughters’ and granddaughters’ reproductive organs.
Studies point to changes in the epigenetic markers related to the development of reproductive organs, which the mothers passed down to their daughters. This finding affirms the adage that “you are what your mother — or grandmother — ate.”
Tagalongs to Our Genetic Code
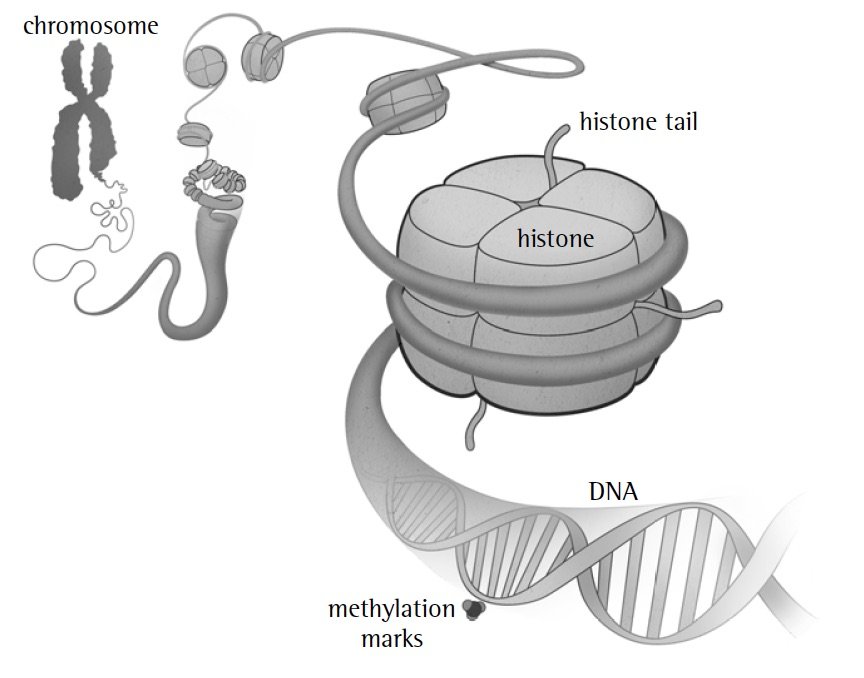
Our DNA includes additional components, which may sometimes be passed from parent to child at the same time as the genetic code. First are molecules attached to the DNA, called methylation marks, that turn genes on and off. Second are balls of proteins composed of histones, which the DNA wraps around. Histones and a portion of these proteins, called histone tails, regulate how the DNA is folded (and thus what is turned on or off).
The food you eat and other aspects of your environment can change these tagalongs. Then they can be passed down to your children and even your grandchildren, affecting the genes that are turned on.
Epigenetics: A Problem for Evolution?
Until these findings, many evolutionists dismissed the ideas of Charles Darwin’s contemporary, Jean-Baptiste Lamarck, who believed that animals could acquire new traits through interactions with their environment and then pass them to the next generation. For instance, he believed giraffes stretching their necks to reach leaves on trees in one generation would cause giraffes in the next generation to have longer necks. Many science textbooks today reject Lamarck’s ideas, but epigenetics is a form of Lamarckianism.
Of course this is contrary to classic Darwinian evolution. The theory of evolution is based on random changes or mutations occurring in DNA. If a change happens to be beneficial, then the organism will survive via natural selection and pass this trait to its descendants.
Although evolutionists do not deny the reality of epigenetics, its existence is hard to explain! Epigenetic changes are not random; they occur in response to the environment via complex mechanisms already in place to foster these changes.
These non-random epigenetic changes imply that evolution has a “mind.” Creatures appear to have complex mechanisms to make epigenetic changes that allow them to adapt to future environmental challenges. But where did this forward-thinking design come from? Evolution is mindless; it cannot see the future. So how could it evolve mechanisms to prepare for the future?
But God does! God is omniscient (all-knowing), and He foreknew Adam and Eve would sin. He would judge that sin (Gen. 3) and the world would be cursed (Rom. 8:22). God knew that organisms would need the ability to adapt in a world that was no longer “very good.” God likely designed organisms with epigenetic mechanisms to allow them to change easily and quickly in relation to their environment. These types of changes are much more valuable than random mutation and natural selection because they can produce immediate benefits for offspring without harming the basic information in the actual sequence of DNA.
Although we often hear that “nothing in biology makes sense except in the light of evolution,” it should be said that “nothing in biology makes sense without the Creator God.” Epigenetics is an exciting field of science that displays the intelligence and providence of God to help organisms adapt and survive in a fallen world.
The New Answers Book 4
Building on the previous New Answers Books, learn more about the Gospel and a young earth, death of plants and leaves, dragons, religious wars, cavemen, science, living fossils, and more.
Read Online Buy BookFootnotes
- Bob Holmes, “Bacteria Make Major Evolutionary Shift in the Lab,” New Scientist (June 2008), http://www.newscientist.com/article/dn14094-bacteria-make-major-evolutionary-shift-in-the-lab.html .
- Kevin L. Anderson and Georgia Purdom, “A Creationist Perspective of Beneficial Mutations in Bacteria,” in the Proceedings of the Sixth International Conference on Creationism (Pittsburgh, PA: Creation Science Fellowship, 2008): p. 73–86.
- Nicola J. Nadeau and Chris D. Jiggins, “A Golden Age for Evolutionary Genetics? Genomic Studies of Adaptation in Natural Populations,” Trends in Genetics 26 (2010): p. 484–492.
- Ibid.
- Ibid.
- Georgia Purdom, “Is Natural Selection the Same Thing as Evolution?” in The New Answers Book 1, ed. Ken Ham (Green Forest, AR: Master Books, 2006), p. 271–282.
- William G. Glass et al., “CCR5 Deficiency Increases Risk of Symptomatic West Nile Virus Infection,” The Journal of Experimental Medicine 203 (2006): p. 35–40.
- Golo Ahlenstiel, et al., “CC-chemokine Receptor 5 (CCR5) in Hepatitis C- at the Crossroads of the Antiviral Immune Response?” Journal of Antimicrobial Chemotherapy 53 (2004): 895–898.
- Vaughn S. Cooper et al., “Mechanisms Causing Rapid and Parallel Losses of Ribose Catabolism in Evolving Populations of E. coli B,” Journal of Bacteriology 183 (2001): 2834–2841.
- Paul Sniegowski et al., “Evolution of High Mutation Rates in Experimental Populations of E. coli,” Nature 387 (1997): 703–705.
- Tim F. Cooper et al., “Parallel Changes in Gene Expression after 20,000 Generations of Evolution in E. coli,” PNAS 100 (2003): 1072–1077.
- Holmes, “Bacteria Make Major Evolutionary Shift in the Lab.”
- Zachary Blount et al., “Historical Contingency and the Evolution of a Key Innovation in an Experimental Population of Escherichia coli,” PNAS 105 (2008): 7899–7906.
- Zachary Blount et al., “Genomic Analysis of a Key Innovation in an Experimental Escherichia coli Population,” Nature 489 (2012): 513–518.
- Blount et al., “Historical Contingency and the Evolution of a Key Innovation in an Experimental Population of Escherichia coli,” 7899–7906.
- William M. Thwaites, “New Proteins without God’s Help,” Creation Evolution Journal 5 (1985): 1–3.
- Ibid.
- Seiji Negoro et al., “Nylon-oligomer Degrading Enzyme/Substrate Complex: Catalytic Mechanism of 6-aminohexanoate-dimer hydrolase,” Journal of Molecular Biology 370 (2007): 142–156.
- Georgia Purdom and Kevin L. Anderson, “Analysis of Barry Hall’s Research of the E. coli ebg Operon,” in the Proceedings of the Sixth International Conference on Creationism (Pittsburgh, PA: Creation Science Fellowship, 2008): p. 149–163.
- Barry G. Hall, “Evolutionary Potential of the ebgA Gene,” Molecular Biology and Evolution 12 (1995): 514–517.
- Ibid.
- Welkin E. Johnson and Sara L. Sawyer, “Molecular Evolution of the Antiretroviral TRIM5 Gene,” Immunogenetics 61 (2009): 163–176.
- Sébastien Nisole, “A Trim5-cyclophilin A Fusion Protein Found in Owl Monkey Kidney Cells Can Restrict HIV-1,” PNAS 101 (2004): 13324–13328.
- Cheng-Hong Liao et al., “A Novel Fusion Gene, TRIM5-cyclophilin A in the Pig-tailed Macaque Determines Its Susceptibility to HIV-1 Infection, AIDS 21 (2007): S19–S26.
- Michael Le Page, “Evolution Myths: Mutations Can only Destroy Information,” New Scientist, April 2008, https://www.newscientist.com/article/dn13673-evolution-myths-mutations-can-only-destroy-information/.
- Terry Devitt, “A Gene Divided Reveals Details of Natural Selection,” University of Wisconsin-Madison News, October 10, 2007, http://www.news.wisc.edu/14276.
- John E. Schienman, et al., “Duplication and Divergence of 2 Distinct Pancreatic Ribonuclease Genes in Leaf-eating African and Asian Colobine Monkeys,” Molecular Biology and Evolution, 23 (2006): 1465–1479.
- Ibid.
- Jianzhi Zhang, et al., “Adaptive Evolution of a Duplicated Pancreatic Ribonuclease Gene in a Leaf-eating Monkey,” Nature Genetics 30 (2002): p. 411–415.
- Cheng Deng, “Evolution of an Antifreeze Protein by Neofunctionalization under Escape from Adaptive Conflict,” PNAS 107 (2010): 21593–21598.
- Diana Yates, “Researchers Show how One Gene Becomes Two (with Different Functions),” University of Illinois at Urbana-Champaign News Bureau, January 12, 2011, http://www.news.illinois.edu/news/11/0112genes_cheng.html.
- “Mutations Are Random,” Evolution 101, https://evolution.berkeley.edu/evo101/IIIC1aRandom.shtml.
- Tom Hennigan, Georgia Purdom, and Todd Charles Wood, “Creation’s Hidden Potential,” Answers, January–March 2009, p. 70–75.
- Jean K. Lightner et al., “Determining the Ark Kinds,” Answers Research Journal 4 (2011): 195–201.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- © 2024 Answers in Genesis




