
Shining Light on the Evolution of Photosynthesis
Originally published in Journal of Creation 17, no 3 (December 2003): 74-84.
Abstract
Biochemical sequences necessary for the evolution of photosynthesis would have required the evolution of a set of sophisticated enzymes that generated a series of useless intermediates.
Summary
Biochemical sequences necessary for the evolution of photosynthesis would have required the evolution of a set of sophisticated enzymes that generated a series of useless intermediates. In the series of enzymes necessary for the manufacture of chlorophyll, these intermediates would have been lethal to the cell before the next enzyme in the series evolved to pick up and modify phototoxic material and insert it into apoproteins. Evidence is presented that: a) the appearance of ground state oxygen would have been lethal to the cell well before oxygen-disarming complexes evolved; b) probability would have eliminated any chance for the evolution of genes for complex enzymes from analogous proteins; c) any junk protein production would have been a death sentence; d) the ATP* synthase motor could not possibly have evolved in a stepwise fashion, and e) the rubisco complex could not and would not have evolved.
* Items marked with an asterisk, the first time they are mentioned, are defined in a glossary at the end of the article.
If we define science as the assumption of naturalism, that matter and energy are all that ever has been or will be, then photosynthesis must of course have evolved, since the only reasonable alternative, that it was designed by intelligence, falls outside our definition of ‘science’. So, by definition, the fabulously complex systems of photosynthesis have arisen by accident. But man’s way of defining words has no jurisdiction in the dominion of objective reality. Truth is sublimely indifferent to our definition of words, even to our definition of science. The thesis of this paper is that evolution would not have been capable of generating the process of photosynthesis as it exists in cyanobacteria, green algae and higher plants, and that it must therefore have been intelligently designed. (For those who are familiar with the intricacies of the photosynthetic system and The Calvin Cycle, please jump ahead to Problem areas for evolution.)
How photosynthesis works: the basics
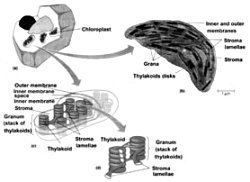
Figure 1. The chloroplast. (a) A cutaway view of a plant cell showing the relative size and orientation of the chloroplasts. (b) A chloroplast as seen by electron microscopy (TEM). (c) A schematic illustration of chloroplast structure. (d) A cutaway of a granum. Figure modified slightly from Becker, W.M., Kleinsmith, L.J. and Hardin, J., The World of the Cell, 4th ed., Chapter 4, p. 91.64 Copyright © 2000 by Addison Wesley Langman Inc., reprinted by permission of Pearson Education Inc.
Eukaryotic photosynthetic organisms have their photosynthetic mechanism isolated in organelles called chloroplasts (Figure 1). A Chlorella green algae cell has one large chloroplast. A modern leaf with 70 million cells will contain about five billion chloroplasts, each containing about 600 million molecules of chlorophyll. Generally speaking, about 250 to 300 chlorophyll molecules per chloroplast will be involved in the transfer of absorbed light energy through neighbouring pigments to a ‘special pair’ of chlorophylls in the reaction centre of their particular photosystem*.1
The chloroplast is surrounded (usually) by two lipid, that is fatty, bilayers. In the green algae and higher plants, chlorophyll is bound to proteins on the internal membranes of the chloroplasts, called lamellae, which are the site of the light reactions* of photosynthesis. The carbon reduction reactions, which ultimately produce sugar from CO2, are catalyzed by water-soluble enzymes that are found in the liquid called stroma, the region of the chloroplast outside the thylakoids.2 Most of the internal chloroplast membranes are closely associated with each other in what are called grana lamellae, which are membrane pouches (thylakoids) stacked like coins, while some membrane sacks are in isolated strands out in the stroma, called stroma lamellae (Figure 1). The parts of the photosynthetic proteins that are inserted in the thylakoid membranes must be mostly hydrophobic (repelling water); the parts that protrude into the lumen* or out into the stroma need to be generally hydrophilic (attracting water).
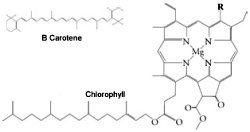
Figure 2. Chlorophylls a and b (right) and B Carotene (above). In chlorophyll a, (R=CH3), and in chlorophyll b, (R=CHO).
Photosynthetic bacteria and eukaryotes collect light energy for photosynthesis in structures called light-harvesting antenna complexes. There are two kinds of reaction centres (RCII and RCI) in the photosystems of eukaryotic photosynthetic organisms that contain chlorophyll a and b, and each has its own kinds of antenna protein complexes. The geometry of the light-harvesting complex proteins of photosystem II (PSII) has been determined. The complex is a transmembrane pigment protein complex, with three α helices* that cross the membrane. About 15 chlorophyll a and b molecules are associated with the complex, along with several carotenoids (see Figure 2). The two long carotenoids form an X in the centre of the complex.3 When too much energy is coming through PSII (the first photosystem to be utilized) for PSI to handle, LHCII complexes can migrate in the membrane to the aid of PSI.4 These antenna complexes are clustered around the reaction centres in the thylakoid membranes and transfer electron excitation energy to the centre complex. The antenna complexes of reaction centre I (RCI) are actually part of the reaction centre complex.5
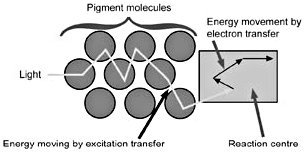
Figure 3. Energy movement within the antenna complex is by resonance transfer, a purely physical process, to the ‘special pair’ of chlorophyll molecules in the reaction centre, where the energy captured is used in the transfer of electrons from chlorophyll to pheophytin, and on to plastoquinones A and B. The original electron source in higher plants is water.
Light energy is captured by pigment molecules in the antenna complexes when photons strike electrons in their conjugated double bonds (alternating single and double bonds). Certain pigments absorb photons at just certain wavelengths. Pigment molecules in antenna complexes, as a general rule, pass their energy along to pigments absorbing at a somewhat lower energy (longer wavelengths) level at each step. Carotenoid pigments can pass their excitation energy to chlorophyll b, which passes its energy to chlorophyll a, which absorbs maximally at 670 nm, and then to the first reaction centre (RCII) pair of chlorophylls, which absorb at 680 nm. The system captures between 95 and 99% of photon energy for use in photochemistry (electron transfer, see Figure 3), sacrificing a tiny amount of energy at each step so that energy transfer is irreversible.6
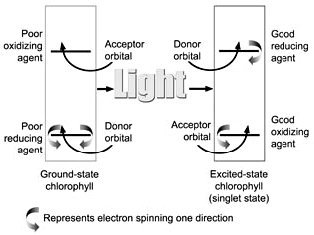
Figure 4. Diagram of orbital occupation for the ground and excited (singlet) states of reaction centre chlorophyll. Arcing arrows stand for electrons of opposite spin direction. In the ground state, the chlorophyll is a poor reducing agent because it can only lose an electron from a low energy orbital, and a poor oxidizing agent because it can only accept electrons in a high energy orbital. In the excited state, an electron can be lost from a high energy orbital, and the molecule becomes an extremely powerful reducing agent. This means that the P680 and P700 excited state molecules have a very negative redox potential.* They ‘want’ to give away electrons very badly.
When a photon is absorbed by a pigment molecule, an electron jumps up from a lower energy orbital, where it is paired with another electron of opposite spin, to a higher energy orbital, leaving both electrons unpaired7 (see Figure 4). This excitation energy is not passed along to the reaction centre by electron transfer, but rather, we think, by resonance transfer,8 like the energy transfer from one tuning fork to another when one is struck and properly placed near the other. This energy transfer is purely a physical process, whereas electron transfer involves chemical changes in molecules.
The last step of the sequence is the excitation of an electron in one of a special pair of chlorophylls (P680) in reaction centre II. The excited electron from this pair is transferred to a molecule called pheophytin, which is a chlorophyll-like molecule in which two protons replace the magnesium in the centre of the (modified) tetrapyrrole molecule. The lightning-fast reactions that follow the reduction of P680 in RCII prevent the loss of the energy harnessed as heat. The electron lost to pheophytin is replaced in the P680 chlorophyll pair from a tyrosine residue (Y) in the reaction centre protein, which in turn derives it from water in a system of four precisely arranged manganese atoms, which are successively oxidized to higher and higher states by light in a process not well understood.9 This system becomes the most powerful oxidizing agent in any known biological system, capable of splitting water molecules to draw off electrons.10
Pheophytin transfers its newly acquired electron to a plastoquinone and this electron is quickly passed to a second plastoquinone (Figure 5). The second quinone picks up two protons from the stroma after it has received two electrons in succession from the first, and then dissociates and carries the electrons to the cytochrome b6f complex where it is oxidized, dumping its two protons into the lumen of the thylakoid and passing its electrons to the cytochrome complex. One of its two electrons is passed through an iron sulfur protein to a cytochrome f and to plastocyanin, on the lumen side, as shown in Figure 3, and one is passed through two cytochrome b molecules to re-reduce an oxidized plastoquinone. This plastoquinone picks up two more protons from the stroma and the process is repeated with two more protons being transferred from the stroma to the lumen. The electron coming through the Rieske iron-sulfur protein and cytochrome f is ferried by plastocyanin to photosystem I (PSI), where it reduces the special chlorophyll pair (P700) of reaction centre I (RCI). This new electron is re-energized by light energy in RCI, and passes to a chlorophyll a (labelled A0 in Figure 611), then to a vitamin K1 (a quinone, labelled A1), and then to a series of iron-sulfur-containing protein complexes to ferredoxin, which can reduce NADP+ to NADPH* via ferredoxin-NADP reductase.12
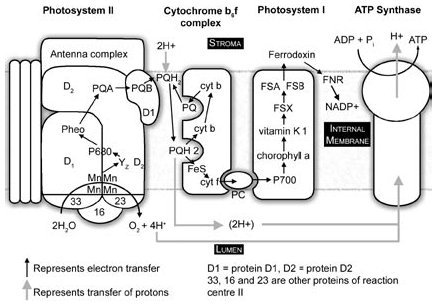
Figure 5. Linear electron transport system of photosynthesis. Photosystem II simplified from Hankamer et al.,65 © 1997 by Annual Reviews <www.annualreviews.org>. Plastocyanin dissociation and movement simplified, because PSI is predominantly in the stroma lamellae.
PSII produces extremely strong oxidizing agents that can pull electrons out of water, but it is not capable of reducing NADP+ to NADPH. PSI produces extremely strong reducing agents that ultimately do the job of reducing ferredoxin and NADP+. Neither system does anything meaningful apart from the other, which is to say, nothing works unless everything works. The whole sequence of events, labelled the Z scheme by Hill and Bendall13 (Figure 6), results in the production of the high energy NADPH molecule. This energy can be utilized in the reduction and regeneration reactions of the Calvin cycle (Figure 7), where CO2 is incorporated into organic molecules and built into triose (three carbon) sugars.
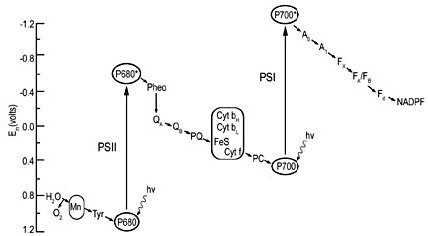
Figure 6. The Z scheme, showing the electron transfer system in terms of redox potentials. From Photosynthesis: A Comprehensive Treatise; reprinted with the permission of Cambridge University Press, and the kind permission of Dr J. Whitmarsh.66
The other energy-accumulating activity resulting from these processes is the accumulation of protons in the lumen of the thylakoids. These protons accumulate as water is split in PSII, and the protons are left in the lumen, and as hydroquinone (the reduced plastoquinone) is oxidized at the cytochrome b6f complex. Protons accumulating in the lumen are channelled back into the stroma through an enzyme complex called ATP synthase (see Figure 8). The CF0 complex of ATP synthase forms a channel through the membrane. The binding sites for ADP and inorganic phosphate and for ATP appear to be on the 3α and 3β subunits of the CF1 complex, alternating like slices of an orange. Evidence seems to support a model in which the asymmetric χ protein rotates within the αβ subunits from the energy of protons entering and exiting at nonaligned sites in the CF0 complex. It appears that the configuration of the β subunit changes so that ADP and inorganic phosphate are bound, then the subunits close, binding the substrates together so that ATP is formed, and then the subunits open out again and ATP is released.14
One ATP is generated for about every four protons that pass through the complex15 ATP and NADPH generated by the light reactions are utilized in the reduction reactions of the Calvin cycle.
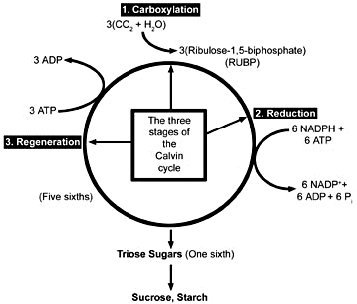
Figure 7. Three stages of the Calvin cycle. 1) In the carboxylation reaction, CO2 is covalently linked to the number two carbon of Ribulose-1,5-bisphate (RUBP). 2) In the reduction reactions, carbohydrate is formed from the 3-phosphoglycerate (PGA) molecules using the energy of ATP and NADPH. 3) In the regeneration reactions, five sixths of the triose sugars are recombined in the simplest possible sets to reproduce RUBP. Two molecules of NADPH and two of ATP are used in the reduction of 3-phosphoglycerate for every CO2 fixed, and one ATP is used in the regeneration reactions for a total of 2 NADPH and 3 ATP’s per carbon fixed.
The Calvin cycle
The Carbon Fixation Reactions (Calvin cycle), the reactions that utilize the energy stored in ATP and NADPH produced in the light reactions to synthesize three-carbon (triose) sugars, (see Figure 7) include three processes:
- Carboxylation of ribulose-1,5-bisphosphate (RUBP), which then divides into two molecules of 3-phosphoglycerate (PGA).
- Reduction of the PGA to triose sugars, consuming 2 molecules of NADPH and two molecules of ATP for each carbon dioxide molecule incorporated, and
- Regeneration of ribulose-1,5-bisphosphate, which consumes one more ATP for each carbon incorporated.16
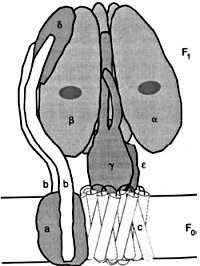
Figure 8. ATP synthase, with F0, membrane-intrinsic, ion-conducting portion, and F1 portion, where ATP is formed, also divided into a rotor portion, comprising c12, ε, and γ polypeptides, and a stator portion, consisting of subunits a, b, δ and (αβ)3. Reprinted from: Junge, W., Lil, H. and Engelbrecht, S., Trends in Biochemical Science 22:420, 1997;50 with permission from Elsevier Science. See ATP synthase is an irreducibly complex motor below.
Figure 9 shows the reactions of the Calvin cycle in greater detail.
The first reaction in Figure 9 is the critical reaction catalyzed by ribulose-1,5-bisphosphate carboxylase/oxygenase, the enzyme we call rubisco. Inorganic CO2 is covalently attached to the second carbon atom of ribulose-1,5-bisphosphate (RUBP). The reaction takes place without ATP, and is actually complex, involving five steps.17 The end products are two 3-phosphoglycerate (PGA) molecules. These molecules are phosphorylated by phosphoglycerate kinase, using ATP, to generate 1,3-bisphosphoglycerate. NADPH is used by the enzyme NADP-glyceraldehyde-3-phosphate dehydrogenase acting on 1,3-bisphosphoglycerate, to produce glyceraldehyde-3-phosphate, the first triose sugar to be generated.18
One sixth of this triose phosphate is used either to produce starch in the chloroplast, or is exported from the chloroplast to be used in the production of sucrose. The remaining five sixths is used in regenerating the molecule (RUBP) the plant uses to capture CO2. What we see in these regeneration reactions is the simplest way of generating sets of five from sets of three, keeping the number of carbon atoms in each molecule as small as possible, except that one-carbon compounds are not allowed.19
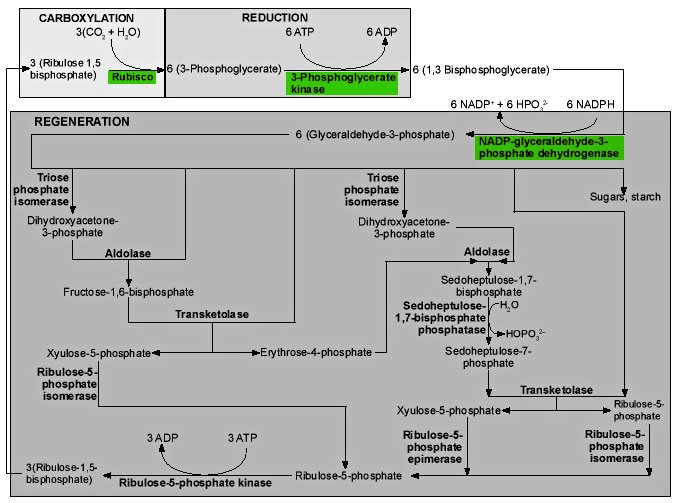
Figure 9. Enzymes (bold type) and reactions of the Calvin cycle. Click thumbnail to view full-size chart.
The absence of any one of the three enzymes, in green in Figure 9, would destroy the balance of the system but theoretically permit the regeneration of some RUBP. In reality, the operation of the CO2 fixation cycle is suppressed if even one of the participating enzymes is inactivated, so that CO2 fixation is lost at low concentrations of H2O2, with a 50% inhibition at only 10μM. If the scavenging system, which takes out this sort of highly reactive molecule, is not functioning, H2O2 can accumulate to this level in 0.5 seconds.20
The last reaction at the bottom of the diagram is also unique to photosynthesis, the utilization of the enzyme ribulose-5-phosphate kinase to add a second phosphate group to ribulose-5-phosphate, generating the critical molecule in the process, RUBP, the substrate for rubisco.
Problem areas for evolution
The assembly of complex molecules involves a series of enzymes that must react in a proper sequence, very often producing intermediates that are useless to the cell until the final product is formed. Evolutionists imagine that these enzymes evolve randomly, often from a duplicate gene, and that the succession of steps in the synthesis, at least often, represents the succession of steps in the historical evolution of the process (the Granick hypothesis). But forces of natural selection could not operate to favour an organism which had ‘evolved’ a series of enzymes which merely produced useless intermediates until it somehow got around to making the end product. The Calvin cycle requires eleven different enzymes, all of which are coded by nuclear DNA and targeted precisely to the chloroplast, where the coding sequence is clipped off at just the right place by a nuclear-encoded protease. In reality, as described in the preceding paragraph, none of the enzymes can be missing if the Calvin cycle is to function. It is true that many of these enzymes are ubiquitous in living systems because every living cell needs to generate ribulose phosphates for the production of RNA, but evolutionists cannot solve the problem by merely pushing it back in time.
The assembly of chlorophyll takes seventeen enzymes.21 Natural selection could not operate to favour a system with anything less than all seventeen being present and functioning. What evolutionary process could possibly produce complex sophisticated enzymes that generate nothing useful until the whole process is complete? Some evolutionists argue that the assumed primeval organic soup had many of the simpler chemicals, and that only as they were used up did it become necessary to generate the earlier enzymes in the pathway. In The Mystery of Life’s Origin: Reassessing Current Theories, the authors set forth the good basic chemistry that demonstrates that there could never have been an organic soup, and present some of the evidence out there in the world indicating that there never was.22 Denton23 and Overman24 also cite a number of experts who suggest that there is no evidence for such a primitive soup but rather considerable evidence against it.
Chlorophyll itself, and many of the intermediates along its pathway of synthesis can form triplet states, which would destroy surrounding lipids by a free radical cascade apart from the context of the enzymes that manufacture them and the apoproteins into which they are inserted at the conclusion of their synthesis.25 According to Asada26 ‘triplet excited pigments are physiologically equivalent to the active oxygens’, and according to Sandmann and Scheer, chlorophyll triplets ‘are already highly toxic by themselves . . . .’27 The entire process of chlorophyll synthesis from δ-aminolevulinic acid to protoporphyrin IX is apparently tightly coupled to avoid leakage of intermediates.28 Almost all of the enzymes of chlorophyll biosynthesis are involved in handling phototoxic material.29 For many of these enzymes, if they are not there when their substrate is manufactured, the cell will be destroyed by their substrate on the loose in the wrong place at the wrong time. Apel30 has cited four of the enzymes of chlorophyll biosynthesis for which this has been proven to be the case. This is a significant problem for evolutionists, who need time for these enzymes to evolve successively. Each time a new enzyme evolved it would have produced a new phototoxin until the next enzyme evolved.
Triplet state chlorophyll, generated in the reaction centres when singlet (excited state) chlorophyll cannot get rid of its energy quickly enough, as may be the case when excess photon energy is coming in, lasts long enough to generate very damaging singlet oxygen (1O2), which attacks lipids, proteins, chlorophyll and DNA.31 Evolutionists maintain that ground-state oxygen (3O2, a triplet state biradical) was not around when photosynthesis evolved. There is, however, considerable evidence that there has never been a time in Earth’s history when there was not significant free oxygen in the atmosphere (see Dimroth and Kimberley,32 Thaxton, Bradley and Olsen,33 Overman and Pannenberg,34 Denton35). The evolutionists’ own analyses suggest that the last common ancestor for the bacteria and archaea already had sophisticated enzyme systems for using O2 and for disarming its reactive by-products.36 Since these organisms had already evolved by 3.5 Ga, on the evolutionists’ timescale,37 this also suggests something rather ominous for the absence of oxygen theory.
In the system that presently exists, a sophisticated complex of enzymes and pigments quenches the excess energy and scavenges the dangerous oxygen species generated by excess light. CuZn superoxide dismutase (in most higher plants) converts superoxide (O2-), the primary product of photoreduction of dioxygen in PSI,38 to H2O2 in the highest-known diffusion-controlled rate among enzymatic reactions.39 It appears that about one molecule of superoxide dismutase attaches to the surface of the membrane in the vicinity of the PSI complex, along with ascorbate peroxidase (APX). Ascorbate reduces the H2O2 generated, in a reaction catalyzed by APX. The product of this reaction, the monodehydroascorbate radical, is reduced again to ascorbate by photoreduced ferredoxin (Fd) in PSI.40 The enzymes and other reducing species of this system could not evolve gradually and then microcompartmentalize over time because nothing works unless everything is in place. This means that the first appearance of oxygen would have been lethal to the cell, whether the source of oxygen was biological or non-biological. Enzymes such as superoxide dismutase would not have been able to evolve at all. APX, for example, has only about 31–33% homology with cytochrome c peroxidase, from which it is thought to have evolved.41 Cells without these enzymes exposed to ground-state oxygen would simply have been destroyed before hundreds of base pair changes generated the enzymes from something else.
Natural selection is not evolution’s friend. In answer to the question, ‘Why would evolution produce a series of enzymes that only generate useless intermediates until all of the enzymes needed for the end product have evolved?’ the evolutionist might respond, ‘Why not?’ The ‘why not’ is in the maths.
The probability of fixation when selection and genetic drift* interact (based on Futyuma42)
The coefficient of selection(s) measures the intensity of selection against less fit genotypes or in favour of more fit genotypes.
If the fitness of genotypes A1A1, A1A2 and A2A2 are 1, 1+ s, and 1+ 2s respectively, where s is positive if A2 is advantageous and negative if it is deleterious, and the initial frequency of A2 is q, the probability of fixation (the new gene replacing the old) of A2 is
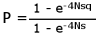
where:
N = effective population size
e = the base of natural logarithms = 2.718If A2 is a new mutation represented by a single copy in the population, q = 1/(2N), and the fixation probability becomes
P = 1/(2N) if s = 0,
which is to say that the probability of fixation is equal to the frequency of the gene in the population if the mutation is neutral. A neutral mutation arising in a (breeding) population of 1,000 individuals would have a probability of 1 in 2,000 (0.0005) of replacing the original gene.
If A2 arises in a population of N = 1,000 individuals, and A2 has a selective advantage of 0.01, the probability of eventual fixation, that is, of A2 replacing A1 in the population, is 0.02 or 1 in 50. A slightly advantageous gene might survive random effects, but probably will not.
If A2 is deleterious, bad, and the selection coefficient against it is only -0.001, the probability of fixation becomes 0.00004 or 1 in 25,000.43 But if s = - 0.01, and N = 1,000, the probability of fixation for this very slightly deleterious gene appearing in a given cell is a little less than one in ten quintillion. Cells producing useless proteins would certainly have been weeded out of existence.
So the answer to the question ‘Why not?’ when we ask the question ‘Why would evolution produce cells or plants with useless proteins until it finally got to the last one and made something useful?’ is that natural selection would have become the ruthless enemy of, rather than the explanation for, the series of events required.
This argument goes in addition to the argument already well known among creationists, that the statistical probability of even 1,000 base pairs aligning correctly to produce a small protein, about 10-600, could not be faced if groups of 1,000 recombined at a rate of a billion per second (109 tries) for 30 billion years (1018 seconds), with the number of bases being equal to the number of electrons that could fit with no space between them into a universe of 5-billion-year radius (10130). This would yield 10157 total tries, an inconceivably huge number, utterly and absolutely trivial in comparison with the number of tries needed to have any chance of generating one small gene.44
The question, ‘Why and how would evolution go about trying to produce a protein for binding pigment molecules before pigment molecules existed?’ is another major challenge for evolutionists.
If chlorophyll evolved before the antenna proteins that bind it, it would in all likelihood destroy the cell, so the proteins had to evolve first. But natural selection could not favour a ‘newly evolved’ protein which could bind chlorophyll and other pigment molecules before those crucial pigments had themselves come into existence! Each binding site must be engineered to bind chlorophyll a or chlorophyll b only or carotene only. The carotene molecules must be present in just the right places for quenching triplet states in the chlorophylls. Even if the pigment molecules were already around, producing just the right protein would be an extremely difficult task. It would not only have to bind pigment molecules only, but it would need to bind just the right pigments in just the right places in just the right orientation so that energy could be transferred perfectly between them, with a little lower energy at each step. Anything else would do nothing, or would transfer energy at random, and the complex would accomplish nothing at best and burn up the cell at worst.
And there is another problem for evolution. The insertion of the pigment molecules changes the conformation of the apoprotein from about 20% to about 60% α-helical content.45 So evolution would have to produce a protein with a wrong shape that would assume just the right shape by the insertion of pigment molecules in just the right positions and orientations when those pigment molecules had not yet evolved.
The energy transfer timeframe between pigment molecules in the antenna complex is between 10-15 and 10-9 seconds. The system that God engineered captures 95–99% of the photon energy for photochemistry, even though there are four other ways the energy can be lost during the slightly less than a billionth of a second the system has for capturing it.46 Humans certainly cannot begin to design systems with such efficiency, but the evolutionists are determined that chance, what Cairns-Smith47 calls ‘old fumble fingers’, can.
Our understanding of the assembly of apoproteins with their pigments is very poor, but we do know that the chloroplast encoded chlorophyll a binding proteins of PSI and PSII core complexes are inserted cotranslationally into the thylakoid. Protein intermediates of the D1 protein have been observed due to ribosome pausing. It may be that this ribosome pausing permits cotranslational binding of chlorophyll a to the protein.48 This kind of controlled insertion, with synthesis of otherwise phototoxic material, is precisely what we would expect from intelligent planning and forethought, but how might ‘old fumble fingers’47 hit on such a scheme?
-
All of the parts must be shipped to the right location, and all must be the right size and shape, down to the very tiniest detail.
ATP synthase is an irreducibly complex motor—a proton-driven motor divided into rotor and stator portions as described and illustrated earlier in this paper (Figure 8). Protons can flow freely through the CF0 complex without the CF1 complex, so that if it evolved first, a pH gradient could not have been established within the thylakoids. The δ and critical χ protein subunits of the CF1 complex are synthesized in the cytosol and imported into the chloroplast in everything from Chlorella to Eugenia in the plant kingdom.49 All of the parts must be shipped to the right location, and all must be the right size and shape, down to the very tiniest detail. Using a factory assembly line as an analogy, after all the otherwise useless and meaningless parts have been manufactured in different locations and shipped in to a central location, they are then assembled, and, if all goes as intended, they fit together perfectly to produce something useful. But the whole process has been carefully designed to function in that way. The whole complex must be manufactured and assembled in just one certain way, or nothing works at all. Since nothing works until everything works, there is no series of intermediates that natural selection could have followed gently up the back slope of mount impossible. The little proton-driven motor known as ATP synthase consists of eight different subunits, totalling more than 20 polypeptide* chains, and is an order of magnitude smaller than the bacterial flagellar motor,50 which is equally impossible for evolutionists to explain.
Evolution cannot account for the assembly and activation of rubisco. All attempts to reconstitute a 16-unit rubisco from any source have failed, so the assembly of rubisco must be studied in the chloroplast extracts.51 The eight large (L) subunits of rubisco are coded by the chloroplast DNA, and the eight small (S) subunits by nuclear DNA. The S subunit of rubisco is synthesized on free cytosolic polyribosomes* and maintained even during synthesis in an unfolded state by chaperones* of the Hsp70 class and their protein partners.52 When the small unit is brought to the import complex of the chloroplast, the fourteen-polypeptide chloroplast Cpn60 chaperonin protein associates with IAP100 (protein) of the import complex and can also associate with mature imported small subunits. The chloroplast Cpn60 chaperone is similar to the E. coli GroEl protein.53 After the unfolded precursor protein enters the stromal space, it binds briefly to a stromal Hsp70 chaperone protein and the N terminal targeting sequence is cleaved.54
The large subunits of the rubisco enzyme are produced by the DNA and machinery of the chloroplast itself and stored complexed to a Cpn60 chaperonin.55,56 This chaperone protein keeps the large subunit protein from folding incorrectly, and therefore becoming useless,57 and is also necessary for the proper binding of the eight large subunits; without it they will form a useless clump.58 In many plants, the large subunits are chemically modified by specialized enzymes59 before they bind to the chaperonin protein. There is strong evidence that chloroplast Cpn60, Cpn21 and Hsp70 also participate in the assembly of the sixteen-unit rubisco complex.60 After a soluble L8 core is formed with the assistance of the chaperonin proteins, tetramers (four-part complexes) of small subunits bind to the top and bottom of the complex to form the complete enzyme.61 There are almost certainly other chaperones and chaperone-like polypeptides or lipo-proteins involved that are not yet characterized.
How do evolutionists explain how natural selection would have favoured a protein complex the function of which was to prevent a still-useless rubisco small subunit from folding outside the chloroplast? Before it evolved a way to get the protein inside, there would be no benefit from keeping it unfolded outside. How could blind chance ‘know’ it needed to cause large subunit polypeptides to fold ‘correctly’ and to keep them from clumping? It could not ‘anticipate’ the ‘correct’ conformation before the protein became useful. And evolution would need to be clever indeed to chemically modify something not yet useful so that it could be folded ‘correctly’ when even the ‘correctly’ folded polypeptide would not yet become useful.
Only a designer would know why it would be necessary to produce a specialized protease, target it to the chloroplast, and program it to clip off the targeting sequence of the small subunit at just the right place. And what about the assembly of a collection of meaningless rubisco parts in just one certain way? In order to design a sophisticated set of tools to make something else useful in the future that had, as yet, no function, evolution (as ‘designer’) would have had to have detailed knowledge of the future usefulness of the protein it was so cleverly engineering. If evolution managed to generate any one of these chaperone protein complexes (and it would not), it would still be useless for generating rubisco unless all the other chaperones were also present. Without any one of them, the sixteen-unit complex could not be generated.
But let us assume the impossible, that evolution succeeded in producing the rubisco enzyme complex, and that random chance happened to generate a new, otherwise useless, enzyme to create its substrate, RUBP. The perfect and complete rubisco sixteen-unit protein complex would then bind tightly to RUBP and do nothing.
In the real world, far away from the never-never land of evolution, another enzyme is needed to separate rubisco from RUBP. Once the rubisco complex is produced, a protein activase uses ATP energy to separate it from RUBP, to which it is tightly bound in its inactive (dark conditions) form. Apparently, the hydrolysis of ATP changes the configuration of the activase protein so that it can bind to rubisco and cause it to release its RUBP. The rubisco must then be carbamylated on the ε-NH2 group of just a certain lysine amino acid residue, and then it must pick up a Mg2+ ion on that carbamyl group* to form the active rubisco site.62 The amide group starts out as NH3+, which must become NH2 before the CO2 can be added, and another proton is lost when the COO- actually attaches, so that these steps are stimulated by low H+ concentration and high Mg2+. Light lowers the H+ concentration of the stroma by a process we have discussed, and raises the Mg2+ also. However, no RUBP can be detected in photosynthetic tissue at night, signifying that it is actually phosphoribulokinase that disrupts the cycle at night.63 What all of this implies is that even if evolution managed the impossible task of generating the rubisco enzyme, the entire system as it presently stands would be needed to turn it on in the light and off in the dark.
Conclusion
It defies common sense to imagine that the irreducible complexity of photosynthetic systems would have arisen according to evolutionary theory. Rather, the incredible organization and intricacy evident in photosynthesis—a process man has yet to fully understand, let alone copy—shrieks of having been designed.
Glossary
Alpha (α) helix—amino acids of a polypeptide arranged in a right-handed spiral or helical pattern, stabilized by hydrogen bonds between the backbone amide and carboxyl groups. Return to text.
ATP—Adenosine triphosphate, the power pack of the cell and the molecule the cell most regularly uses to drive uphill, energy requiring reactions. Return to text.
Carbamyl group—a carboxyl group bonded to a nitrogen atom. Return to text.
Chaperone—protein complexes that prevent folding, or prevent improper folding of other polypeptides, or that cause other polypeptides to assume the correct configuration. The twenty-one polypeptide complexes that enclose proteins inside of a ‘can’ and then close a lid at one end to engender proper folding are sometimes referred to as chaperonins. Return to text.
Genetic drift—the effect of random fluctuations on gene frequencies, sometimes resulting in the elimination of one allele (form of a particular gene) from the population. Return to text.
Light reactions—the processes that begin with the excitation of chlorophyll by light and culminate in the synthesis of ATP and NADPH (see Figure 3). Return to text.
Lumen—the liquid chamber inside of a thylakoid, which becomes more acidic in the light. Return to text.
NADPH—nicotinamide adenine dinucleotide phosphate, molecule generated by the electron transport system of the photosynthetic apparatus, used in reduction and regeneration reactions of the Calvin cycle. Return to text.
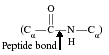 Peptide (amide) bond—the link between the carboxyl (COOH) and amide (NH2) group of two amino acids, formed by the loss of a water molecule (condensation). Return to text.
Peptide (amide) bond—the link between the carboxyl (COOH) and amide (NH2) group of two amino acids, formed by the loss of a water molecule (condensation). Return to text.
Photosystem—the protein and pigment systems of chloroplasts that transform light energy into chemical energy (see Figure 3), including the light harvesting antenna complex and the reaction centre proper, where electron transfer takes place. Return to text.
Polypeptide—a single continuous chain of amino acids bound by peptide (amide) bonds. A protein may have more than one polypeptide. Return to text.
Redox potential—the tendency of a substance to gain or donate electrons, its ‘electron pressure’. Return to text.
Ribosomes—the protein and RNA complexes that are the primary protein-generating factory of the cell. Return to text.
About the Author
Rick Swindell received a Ph.D. in plant breeding and genetics at the University of Missouri Columbia (UMC) in 1976. He taught as a student at UMC, and as an assistant professor at Virginia Tech. Following post-doctoral work in plant breeding at Centro Internacional de Agricultura Tropical (CIAT) in Cali, Colombia, South America, he obtained a M.Div. at Liberty University, Lynchburg, and then went into Christian ministry. Return to top.
Footnotes
- Von Wettstein, D., Gough, S. and Kannangara, C.G., Chlorophyll biosynthesis, The Plant Cell 7:1039–1057, 1995.
- Taiz, L. and Zeiger, E., Plant Physiology, 2nd Edition, Sinauer Associates Inc., Massachusetts, pp. 155–225, 1998.
- Taiz and Zeiger, Ref. 2, p. 172.
- Taiz and Zeiger, Ref. 2, p. 185.
- Nitschke, W., Muhlenhoff, U. and Liebel, U., Evolution; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 285–305, 1998.
- Taiz and Zeiger, Ref. 2, pp. 170–171.
- Taiz and Zeiger, Ref. 2, p. 174.
- Taiz and Zeiger, Ref. 2, p. 170.
- Taiz and Zeiger, Ref 2, pp. 177–178.
- He, W.Z. and Malkin, R., Photosystems I and II; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 29–43, 1998.
- Whitmarsh, J., Electron transport and energy transduction; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 87–107, 1998.
- Taiz and Zeiger, Ref. 2, pp. 178–181.
- Hill, R. and Bendall, F., Function of the two cytochrome components in chloroplasts: a working hypothesis, Nature 186:136–137, 1960.
- Boyer, P.D., The ATP synthase—a splendid molecular machine, Ann. Rev. Biochem. 66:717–749, 1997.
- Taiz and Zeiger, Ref. 2, p. 182.
- Taiz and Zeiger, Ref. 2, pp. 195–225, 1998.
- Sharkey, T.D., Photosynthetic carbon reduction; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 111–122, 1998.
- Taiz and Zeiger, Ref. 2, pp. 198–201.
- Sharkey, Ref. 17, p. 115.
- Asada, K., Radical production and scavenging in the chloroplasts; in: Baker, N.R. (Ed.), Photosynthesis and the Environment, Kluwer Academic Publishers, Dordrecht, Netherlands, pp. 123–150, 1996.
- Beale, S.I., Enzymes of chlorophyll biosynthesis, Photosynthesis Research 60:43–73, 1999.
- Thaxton, C.B., Bradley, W.L. and Olsen, R.L., The Mystery of Life’s Origin: Reassessing Current Theories, Lewis and Stanley, Dallas, pp. 42–68, 1984.
- Denton, M., Evolution: A Theory in Crisis, Adler & Adler, Bethesda, pp. 249–273, 1986.
- Overman, D.L. and Pannenberg, W., A Case Against Accident and Self-Organization, Rowman & Littlefield Inc., Lanham, Boulder, New York, Oxford, pp. 31–67, 1997.
- Apel, K., Metabolism and strategies of higher plants to avoid photooxidative stress, in: Aro, E-M. and Andersson, B. (Eds.), Regulation of Photosynthesis, Kluwer Academic Publishers, Netherlands, pp. 235–252, 2001.
- Asada, Ref. 20, p. 127.
- Sandmann, G. and Scheer, H., Chloroplast pigments: chlorophylls and carotenoids; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 44–57, 1998.
- Sandmann and Scheer, Ref. 27, p. 48.
- Beale, Ref. 21, pp. 45–46.
- Apel, Ref. 25, p. 238.
- Apel, Ref. 25, p. 236.
- Dimroth, E. and Kimberley, M.M., Precambrian atmospheric oxygen: evidence in the sedimentary distributions of carbon, sulfur, uranium and iron, Canadian J. Earth Sciences 13(9):1161–1185, 1976.
- Thaxton et al., Ref. 22, pp. 78–82.
- Overman and Pannenberg, Ref. 24, pp. 41–43.
- Denton, Ref. 23, pp. 261–262.
- Blankenship, R.E., Molecular Mechanisms of Photosynthesis, Blackwell Sciences Ltd., Oxford, pp. 220–257, 2002.
- Blankenship, Ref. 36, pp. 224–225.
- Asada, Ref. 20, p. 130.
- Asada, Ref. 20, p. 131.
- Asada, Ref. 20, p. 143.
- Asada, Ref. 20, p. 135.
- Futyuma, D.J., Evolutionary Biology, 3rd ed., Sinauer Associates Inc., Massachusetts, pp. 365–396, 1998.
- Futyuma, Ref. 42, p. 393.
- Morris, H., Scientific Creationism, 2nd ed., Master Books, El Cajon, pp. 59–90, 1985.
- Chitnis, P.R., Import, assembly and degradation of chloroplast proteins; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 58–71, 1998.
- Taiz and Zeiger, Ref. 2, p. 170.
- Cairns-Smith, A.G., Seven Clues to the Origin of Life: A Scientific Detective Story, Cambridge University Press, Cambridge, London, pp. 98–106, 1985.
- Paulsen, H., Pigment assembly—Transport and ligation; in: Aro E-M. and Andersson, B. (Eds.), Regulation of Photosynthesis, Kluwer Academic Publishers, Netherlands, pp. 219–233, 2001.
- Kapoor, S. and Sigiura, M., Expression and regulation of plastid genes; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 72–86, 1998.
- Junge, W., Lill, H. and Engelbrecht, S., ATP synthase: an electrochemical transducer with rotary mechanics, Trends in Biochem. Sci. 22:420–423, 1997.
- Roy, H. and Andrews, J., Rubisco: assembly and mechanism; in Photosynthesis: Physiology and Metabolism, Advances in Photosynthesis, Vol. 9, Kluwer Academic Publishers, Dordrecht, pp. 53–83, 2000.
- Chitnis, Ref. 45, p. 64.
- Roy and Andrews, Ref. 51, p. 56.
- Lodish, H., Berk, A., Zipursky, S.L., Matsudaira, P., Baltimore, D. and Darnell, J., Protein Sorting: Organelle Biogenesis and Protein Secretion; in: Molecular Cell Biology, 4th Ed., W.H. Freeman and Company, New York, pp. 675–750, 2000.
- Lodish et al., Ref. 54, p. 686.
- Roy and Andrews, Ref. 51, p. 56.
- Lodish, Ref. 54, p. 686.
- Hughes, M.A., Plant Molecular Genetics, Addison Wesley Longman Ltd., Harlow, England, pp. 36–50, 1996.
- Roy and Andrews, Ref. 51, p. 59.
- Roy and Andrews, Ref. 51, p. 57.
- Roy and Andrews, Ref. 51, p. 58.
- Taiz and Zeiger, Ref. 2, p. 206.
- Sharkey, T.D., Photosynthetic carbon reduction; in: Raghavendra, A.S. (Ed.), Photosynthesis: A Comprehensive Treatise, Cambridge University Press, Cambridge, pp. 87–207, 1998.
- Becker, W.M., Kleinsmith, L.J. and Hardin, J., The World of the Cell, 4th ed., pp. 78–109, 2000.
- Hankamer, B., Barber, J. and Boekema, E.J. Structure and membrane organization of photosystem II in green plants, Annual Review of Plant Physiology and Plant Molecular Biology 48:643, 1997.
- Whitmarsh, Ref. 11, p. 91.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- © 2024 Answers in Genesis

