
FOXP2 and the Non-Evolution of Human Language
Even though FOXP2 has a similar sequence and a similar function in other organisms, it serves a unique role in the context of a human being to facilitate language.
As a creation scientist, I have become used to the frequent media hype surrounding “new evidence” that confirms “Darwin was right all along.” Occasionally, I will be asked to investigate and write on these discoveries from a creationist perspective. After reading the original scientific reports or other articles, I usually find a significant difference between the hype and the facts.
A case in point is a recent article in The New York Times1 on FOXP2, a gene that, when mutated, is linked to speech and language impairment. According to the article:
If language had been built on a completely unprecedented set of genes, Darwin (and his successors) would have a lot of explaining to do. With no more than a few hundred thousand years to play with, a linguistic system that depended on thousands of evolutionarily unprecedented genes would seem impossible. But evolution is about random processes that tinker with old parts, not about engineering new ones.
Most of the genes involved in language have some sort of close and ancient counterpart in other species. As a case in point, consider the first gene to be unambiguously tied to language, known as FOXP2, discovered by Simon Fisher and Anthony Monaco, Oxford geneticists.
Rather than emerging from scratch in the course of human evolution, FOXP2 has been evolving for several hundred million years—in a way that placed it perfectly for evolving a critical role in language acquisition.
Thus, it would seem that scientists have found a gene responsible for human language—a gene present in other organisms that only needed to be tweaked a little to allow for the evolution of human language.
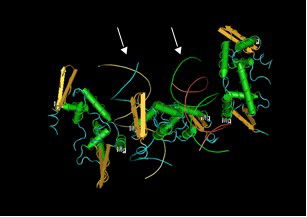
Partial structure of human FOXP2 bound to DNA.2 DNA double helix molecules are indicated with arrows. Transcription factors such as FOXP2 typically bind to specific short sequences of DNA in order to regulate which genes are turned on and which are turned off. Mutation or loss of a transcription factor results in mis-regulation of proteins. The impact of such proteins is the types and amounts of other proteins that are produced.
While many animals do have the FOXP2 gene (including chimpanzees, gorillas, orangutans, mice, birds and even zebrafish!) and mutations in FOXP2 lead to impaired vocalization in other organisms, there is much more to the story.
First of all, the FOXP2 protein is a transcription factor.3 That means it is a DNA-binding protein and serves as a switch controlling the production of other gene products. Therefore, FOXP2 is not directly involved in language. Instead, it is serving as a master switch, controlling the expression of other genes including possibly other transcription factors that promote the development of neurons that will be involved in language.
Such master control genes can be compared to circuit breakers in a fuse box. When switched off, they prevent power from going to several electrical outlets and light switches. Without the circuit breaker on, there will be no light, although the circuit breaker is not sufficient to produce light itself. Likewise, mutations to FOXP2 can have effects on many different products. Although the exact target genes controlled by FOXP2 are unknown, scientists believe that the protein serves as an upstream regulator of development of certain neurons and other tissues. If those neurons do not develop appropriately, then it is possible to have permanent behavioral deficits. In this case, the behavioral deficit is impaired language ability.
Because FOXP2 is a transcription factor, it cannot control language by itself. Obviously, there are many other proteins involved in the process. Indeed, mutations of FOXP2 primarily result in impairment of orofacial movements preventing those affected from making proper word sounds. Afflicted individuals also show some slight grammar deficiencies. Although the gene is clearly linked to language production, one cannot designate it as the “language gene.” An analogy would be calling the mutation that causes muscular dystrophy (and an inability to walk) a “walking gene.”
Researchers have clearly been surprised by the virtually identical sequence of FOXP2 across species. Indeed, FOXP2 is the same in chimpanzees, gorillas, and macaques. There is only one difference in the amino acid sequence between those primates and mice in spite of the fact that they are supposed to be separated by 130 million years of evolution. This is all the more surprising since humans and chimps have two amino acid differences.4 Thus, there are only three out of ~700 amino acids different between humans and mice. No doubt this high degree of similarity is surprising given the clear differences in language usage between humans and mice.
Even with the small difference in amino acid sequence, it does not follow that these differences are responsible for human language ability. We know that FOXP2 is necessary for proper language utilization in man, as demonstrated by the impairment accompanied with FOXP2 mutations. However, FOXP2 is not sufficient for language, otherwise we should expect a mouse or a chimpanzee genetically engineered with human FOXP2 to use human language. Apparently there is much more to human language than just this one gene.
Human language is multi-faceted and involves many different specialized areas of the brain. One recently identified brain region involved in language is the left occipitotemporal cortex.5 Amazingly, this part of the brain recognizes visual word forms. It processes the shape of a written word. Prior to this discovery, a Visual Word Form Area (VWFA) has been controversial because the usage of writing has only a few thousand years of history—long after humans supposedly evolved. In an evolutionary paradigm, selection for the VWFA would have to take place before writing was invented. Thus, it would be difficult to see how such a neuronal system could evolve through natural selection.
Context is a crucial aspect of language and context will help us make sense of the FOXP2 protein. This protein can have a nearly identical sequence and play a similar role6 in different organisms, but this similarity does not prove that language evolved. For example, scientists have been able to transfer these developmental transcription factors from one organism to another. Scientists were able to induce eye development in flies using the similar master control gene from mice!7 As expected, the mouse master control gene induced the production of a typical eye in flies and not a mouse eye. This is because the substituted gene is still in the context of the fly. Even though FOXP2 has a similar sequence and a similar function in other organisms, it serves a unique role in the context of a human being to facilitate language.
Footnotes
- Marcus, Gary, “From Squeak to Syntax: Language’s Incremental Evolution” The New York Times, April 11, 2006.
- Chen, J., Anderson, J.B., DeWeese-Scott, C., Fedorova, N.D., Geer, L.Y., He, S., Hurwitz, D.I., Jackson, J.D., Jacobs, A.R., Lanczycki, C.J., Liebert, C.A., Liu, C., Madej, T., Marchler-Bauer, A., Marchler, G.H., Mazumder, R., Nikolskaya, A.N., Rao, B.S., Panchenko, A.R., Shoemaker, B.A., Simonyan, V., Song, J.S., Thiessen, P.A., Vasudevan, S., Wang, Y., Yamashita, R.A., Yin, J.J., Bryant, S.H., MMDB: Entrez's 3D-structure database, Nucleic Acids Res. 31(1):474-7, January 2003.
Stroud, J.C., Wu, Y., Bates, D.L., Han, A., Nowick, K., Paabo, S., Tong, H., Chen, L., Structure of the forkhead domain of FOXP2 bound to DNA, Structure 14(1):159-166, January 2006. - Enard, W., Przeworski, M., Fisher, S.E., Lai, C.S.L., Wiebe, V., Kitano, T., Monaco, A.P., Paabo, S., Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418:869–872, 2002.
- The two differences in amino acid sequence between humans and apes are especially intriguing. One of the differences is that apes have threonine instead of asparagine at position 303, while humans have serine instead of asparagine at position 325. The reason this is significant is that both of these are potential phosphorylation sites and thus could be ways that the protein is switched on and off.
- Gaillard, R., Naccache, L., Pinel, P., Clemenceau, S., Volle, E., Hasboun, D., Dupont, S., Baulac, M., Dehaene, S., Adam, C., and Cohen, L., Direct intracranial fMRI and lesion evidence for the causal role of left inferotemporal cortex in reading, Neuron 50:191–204, 2006.
- A transcription factor responsible for controlling the development of neuronal populations involved in production of sounds.
- Halder, G., Callaerts, P., Gehring, W.J., Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267:1788–92, 1995.
Recommended Resources

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- © 2025 Answers in Genesis







