The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Jeanson (2017a) takes on a formidable task: To show that the theory of evolution is wrong, and to replace it with biblical creation. To make it short: Jeanson fails.
Let me start by praising Jeanson for his easy-to-read and easy-to-understand style. The book is readable by the broad public, and it has a brilliant account of the basics and history of genetics. All details necessary to understand the arguments are clearly presented.
Biogeography, taxonomy, genetic diversity, and speciation are the main topics, which, according to Jeanson, the theory of evolution either fails to explain or which can be explained without reference to evolution.
I’ll go through the four topics one by one; give an ultrashort account of Jeanson’s argument and explain why I think he fails to prove his point.
Biogeography
This is the study of the distribution of species over the world. Jeanson describes in some detail why he thinks migration from Eurasia to the rest of the world can explain the current distribution of animals. He concludes that “Migration fits the current geographic distribution of species” (Jeanson 2017a).
This is a bold assertion. A few examples will show why.
Of 19 families of marsupials, 17 are endemic to Australia and the nearby islands. (endemic: a species or group of species living exclusively in a well-defined area). Jean Lightner, an associate of the Creation Research Society, has identified all these families as separate biblical kinds in her overview of mammalian ‘Ark kinds’ (Lightner 2012). It is therefore relevant to ask how it comes that all these animals migrated from the Middle East to Australia, leaving no trace behind them, if the biblical story of the Flood is true. Further, they were only followed by those placental mammals that have the best chance of traveling over the sea (a few families of bats and one family of rodents). What a coincidence! I love to think about the poor marsupial mole digging its way from Turkey (Mt. Ararat) to Australia, trying to keep up with kangaroos, koalas, wombats, and numerous crawling, hopping, and gliding marsupials.
In Central and South America, there are four endemic families of monkeys (no family of monkeys live in both Central and South America and other areas). All four are recognized as separate ‘kinds’ by Jean Lightner, who recognizes a total of 15 primate ‘kinds.’ Judged by the homology of the mitochondrial genome (hereafter mtDNA), the Central and South America monkeys are closer related to one another than to other groups. This is reflected in the fact that they are all members of the so-called ‘parvorder’ (a group-level between family and order) Platyrrhini (New World Monkeys). According to evolution, this is because all four originated from a single group of monkeys that made it to South America after it split from Africa by plate tectonics. According to creation, they must have made it to South America separately after the end of the Flood. What a coincidence!
Correspondingly four of five ‘kinds’ of primates on Madagascar are lemurs, which are endemics of Madagascar. What a coincidence!
Conclusion: Jeanson fails to account for biogeography, while the topic is among Darwin’s original arguments in favor of evolution.
Taxonomy
An important difference between the theory of evolution and creationism is the interpretation of the biological taxonomical hierarchy. According to the theory of evolution, all levels of the hierarchy reflect common descent. According to creationism, no level above the created ‘kinds’ (in vertebrates more or less the family-level) reflects common descent.
According to Jeanson (Chapter 5), common descent is not needed to explain the nested hierarchies. He uses the analogy of vehicles to explain how such a nested hierarchy would result from design as well. For example, in vehicles the Family level could be cars, pickups, SUV and the like. The Order level would include tractors, the Class level would include military tanks, and the Phylum level would include airplanes. This Phylum would be ‘Powered Vehicles.’ Another Phylum could be ‘Unpowered Vehicles’ exemplified by a hang-glider. All these belong to the Kingdom of ‘Vehicles.’ Other Kingdoms, would be other devices: computers, washing machines, tools and so on. Jeanson’s corresponding examples in animals are Family: Horses; Order: Odd-toed ungulates (horses, rhinos and tapirs); Class: Mammals; Phylum: Chordates. Kingdom: All animals.
However, in designed objects, many of the smallest parts are exactly the same: the same kind of batteries, wires and LEDs are used in different devices. The same kind of artificial polymers and metal alloys are used as well. Also, more complex parts follow this pattern. The use of diesel engines vs. gasoline engines vs. electrical engines does not follow any reasonable taxonomy of vehicles. The problem goes further than just the common use of these things. One model of airplane, ferry, and car could use an upholstery constructed of, say, 10% rayon and 90% nylon. Another model of airplane, ferry, and car could use an upholstery constructed of, say, 90% rayon and 10% nylon.
When it comes to living organisms, a corresponding violence of the hierarchical pattern would hardly be possible. When you look at, for example, structure of the cell membrane, it follows large taxonomical groups. Any protein sequence from the GenBank database is unique to the species or genus it comes from. The structure of the eye of vertebrates differs from that of mollusks by the arrangement of retina and nerve cells. Arthropods have yet another arrangement. Other arrangements can be found in various other phyla, always following the taxonomical groups.
Why does the skeleton of whale flippers resemble that of land mammals rather than that of shark fins (which have the same overall function as the whale flipper)? All examples where evolution put restrictions on the ‘design.’
Another objection to Jeanson’s model is that there is no ‘natural’ hierarchy of vehicles. His major groups are ‘Powered’ and ‘Non-powered.’ But why not ‘Military’ and ‘Civilian’ (historically that would make more sense), or ‘For transportation of people’ and ‘For transportation of goods’? Or why not use ‘Powered’ vs. ‘Non-powered’ as the main categories, including washing machines in the Kingdom ‘Powered devices,’ while furniture, hand-tools and bicycles is placed in the Kingdom ‘Non-powered devices.’
Contrary to this, evolution immediately suggests a natural hierarchy: that based on descent. Today descent is mostly evaluated by comparing DNA-sequences for at least two reasons: DNA is the ultimate source of variation; and the details of information in DNA is much larger than in any other group of characters.
In practice, it is a problem that not all genes suggest the same phylogeny. This is due mainly to the stochastic process of mutation. Dealing with more genes at the same time (as in mtDNA) mostly solves the problem. To put the argument to the limit, look at a motor glider and a normal glider (aircrafts). They are virtually identical. However, according to Jeanson’s system, the motor glider belongs to the phylum ‘Powered vehicles’ while the normal glider belongs to the phylum ‘Non-powered vehicles,’ and as such, they should be more different than an electrical bicycle and a joint strike fighter (both belonging to the same phylum). What could be more ridiculous? Biological hierarchies, based on evolution, would never end up in such self-contradicting nonsense.
Conclusion: Jeanson fails to account for the nested hierarchy of living organisms, while it is an inevitable part of theory of evolution as it directly reflects common ancestry.
Genetic Diversity
According to Jeanson, most of the genetic variation within ‘kinds’ existed before speciation, and can be explained as original created variation. The theory of evolution agrees that genetic diversity existed prior to speciation but ascribes it to accumulation of mutations over past eons. Jeanson uses great apes as one example among others (Chapter 10). He excludes humans from the family, as humans are not part of the great ape ‘kind.’ Jeanson relies on the nuclear genome in his analysis. However, if you compare homology between nuclear genomes from humans, chimps, gorillas, and orangutans you have no choice. If homology between nuclear genomes reveals relationship, chimps are closer related to humans than to gorillas and orangutans (Prado-Martinez et al. 2013). If homology between nuclear genomes has nothing to do with relationship, Jeanson’s calculations are worthless. What is it?
Though data from the GenBank database could be used to test Jeanson’s idea that the majority of genetic variation within families is due to original created variation, he makes no attempt to do so. To illustrate how this could be done, I have used information about differences in 15 nuclear genes in cats (Johnson et al. 2006). Cats are among the ‘kinds’ that, according to the Bible, were present at the Ark in only one pair. Therefore, a maximum of four alleles1 of each gene could be present in this original pair. This should be traceable in the current variation within the cat family.
Fig. 1 shows the sum of differences found in these 15 genes.
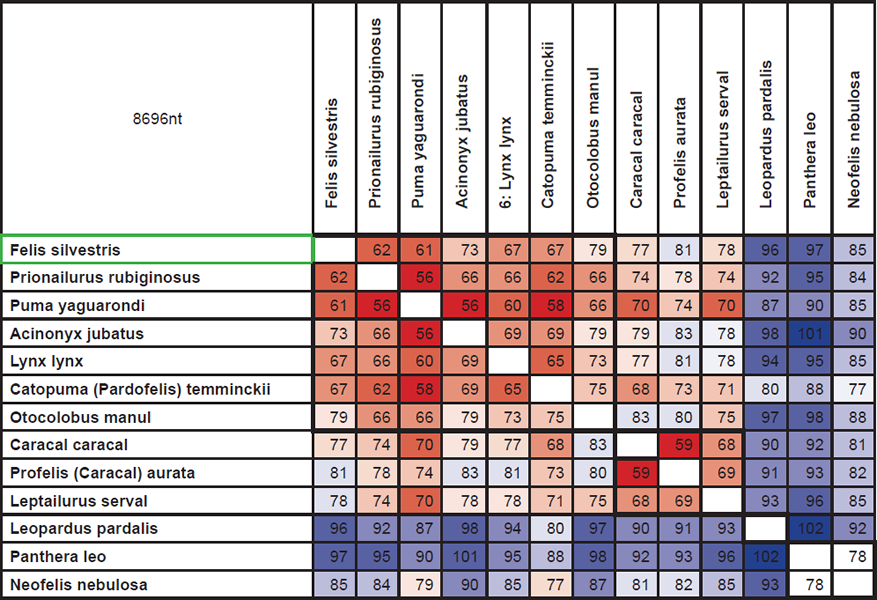
Fig. 1. Numbers are the sum of differences in DNA sequences found in 15 protein coding nuclear genes in various species of cats (the family Felidae). Colors show homology. Low homology: blue. Medium homology: white. High homology: red. 8696nt: The total length of the DNA sequences of the 15 genes in question. Bold lines indicate four species groups, corresponding to four original alleles. See main text for further explanation (Johnson et al. 2006).
To squeeze these results into four groups (the squares) reveals that at least 79 mutations must have occurred. According to creationism 4400 years has passed since the Flood, so there is a maximum of 4400 generations (no cats start breeding before they are two years old on average) between the most distantly related species. 79 mutations in 4400 generations in a sequence of 8696 nt correspond to 1 mutation in 500.000 nt per generation. In humans, according to Jeanson, the corresponding number is 1:40 million (78 mutations per generation per genome, 3.1 billion nt). Cats would have to have an almost 80 times higher mutation rate than humans. At best an extremely bold prediction.
Multifunctionality of mitochondrial genes
A related topic is the fact that whenever genes are present in larger groups of species, such as mammals, all animals, or even all eukaryotes (organisms with a cell nucleus), the homology of the genes resemble the homology of the anatomy. mtDNA has no known function related to anatomy or physiology, yet the homology between mtDNA reflects that of anatomy throughout the biological system. Jeanson’s solution is to ascribe additional functions to the mitochondrial genes (Chapter 7). Such function should discriminate marsupials from other mammals, beetles from butterflies, squids from oysters and all the numerous groups of singled celled eukaryotes from each other. What he actually suggests is several functions of each gene. With no shred of evidence!
I am not sure Jeanson actually has realized what a challenge he has given himself. Let’s take the mitochondrial protein ‘Cytochrome oxidase subunit 1’ or just ‘Cox1’ as an example, starting with the dog family (Canidae).
Jeanson has to suggest, and ultimately identify, one or more function(s) of the Cox1 protein that can explain why this gene can be used to discriminate between Canidae and other families in the suborder Caniformia (such as bears and seals). He has to repeat this process (with either the same or another function) and explain why Cox1 can be used to discriminate between Caniformia and the suborder Feliformia (for example, cats and hyenas). Caniformia and Feliformia are suborders of the carnivore order, Carnivora, and Jeanson has to make the same explanations about the discrimination of Carnivora from other orders of placental mammals. Then the process should be repeated, comparing placentals, marsupials and monotremes, which together form the class Mammals. Next level is to compare Mammals with other classes of tetrapod vertebrates such as birds and crocodiles. Next, the tetrapods should be compared to other vertebrates such as bony fish and sharks. Vertebrates is a subphylum of the phylum Chordata, so vertebrates should be compared to invertebrate subphyla within the chordates, for example, the tunicates. Chordates then should be compared to other phyla, for example, arthropods or mollusks. We could extend the investigation to other kingdoms: plants or fungi, or to the plethora of single celled eukaryotes, who also have mitochondria.
Every single level can be recognized by their Cox1 protein sequence. On every level, homology within the group is as high or higher as it is when species within the group is compared to species outside the group. Jeanson suggests that this complicated pattern reflects function.
Instead of starting with the wolf, we could start with the ladybird (insects), the thale cress (plants), the portobello (fungi) or even the malaria parasite (single celled eukaryote).2 All have their own version of the Cox1 protein. The task of suggesting function to all these levels of living organisms is overwhelming. It is not enough to suggest one specific function. Jeanson still has to explain why the function in question reflect the hierarchical structure of taxonomy. If we, for example, accept that Cox1 has influence on fur-color, why then, is Cox1 from wolf more homologous to that of a leopard, than to that of a horse?
However, Jeanson’s problems don’t stop here. This is just the function of the proteins. The proteins are coded by genes. The majority of differences on the gene level (the DNA-sequences), are so-called silent or synonymous substitutions. These are differences between two protein-coding DNA-sequences that do not result in a difference at the protein level. If creation is true, and the differences between proteins are functional, the synonymous substitution must be functional as well. Otherwise, why would they reflect the taxonomy? Jeanson should suggest what those functions could be and how they can be so important that they outnumber the non-synonymous substitutions.
However, Jeanson’s problems don’t stop here.
About one third of the mtDNA in animals does not encode proteins but are so-called tRNA, rRNA or the D-loop (also called the control region). What anatomically relevant functions can be attributed to these regions?
The function of the various mtDNA sequences is well known. It is therefore Jeanson who has the burden of proof. I challenge him to suggest relevant functions, and explain how they fit the pattern outlined above. Until he does so, I will claim that he cannot.
From an evolutionary point of view, all these patterns of homology simply reflect the distance to the common ancestor.
Selection
Jeanson makes a number of other analyses on the mtDNA. In all cases, he fails to include selection, though this can be shown to be a very real phenomenon. To understand this you have to know that most mutations in protein coding genes fall into two categories: synonymous mutations that do not alter the resulting protein, and non-synonymous that do alter the protein. Due to the way DNA is translated into protein, about 21% of all mutations are synonymous (See Chapter 3, especially Table 3.1 and the appendix). I compared two of the most different human mtDNAs (GenBank numbers EF184607 and FJ168742) and counted the non-synonymous and synonymous differences in the protein-coding genes. 35 of 55 = 64% were synonymous. If more distant mtDNA sequences are compared (Modern Human: KC345974 and Denisovan: FR695060) 85% are synonymous. This tells us that non-synonymous mutations are under stronger selection than synonymous. Selection is thus important, and Jeanson should include it.
Jeanson doesn’t accept the mtDNA sequences from Neanderthals and Denisovans. He claims that the sequences are unreliable, partly because the DNA has been degraded or contaminated. I have urged him to confront the relevant scientists, but he refuses to do so for reasons I don’t think are valid (Frello 2017; Jeanson 2017b). If mtDNA from Denisovans is unreliable, the mistakes in the sequence should be expected to be randomly distributed, when counted as synonymous vs non-synonymous substitutions. A short look at the results mentioned above show that this is far from the case. To dismiss Denisovan mtDNA as unreliable is thus unfounded.
Conclusion
Jeanson fails to account for the pattern of genetic homology. According to the theory of evolution, the homology of both nuclear DNA and mtDNA is correlated with that of anatomy because both are due to the pattern of descent. The inclusion of humans with the other great apes is an integrated part of the theory.
Speciation
Jeanson deals with this in several ways (Chapter 6). First, he notices that the number of breeds of horses, dogs etc. is much larger than the number of species within the relevant family. Jeanson concludes that speciation within a biblical timeframe is unproblematic. He makes no calculations to support his claim. This is rather strange, because a method for doing so is right in front of his eyes: the mtDNA that he uses in other arguments.
As soon as such an analysis is made, the problems appear. A few calculations on mtDNA from dogs will illustrate the problem. Between mtDNA from domesticated dogs and wolves, there are about 170 differences. The dog family falls into two large groups: One includes wolf, dhole and the African wild dog; the other includes various species of foxes.3 The mtDNA from the two groups has between 2400 and 2600 differences between them. Not taking selection and differences in generation time into account, this means that the dog family is at least 15 times older than the domestication of dogs from their wolf ancestors. Taking selection into account will make this difference even larger. It should be noted that in the GenBank, mtDNA are missing from five genera (one of which is extinct) and several species of the dog family. Therefore it might be that the most different mtDNAs is yet to be identified. It also should be noted that the dog family is often used by creationists as an example of a biblical ‘kind’ and acknowledged as such by Lightner in her analysis of mammalian Ark kinds (Lightner 2012, 151–204).
Jeanson further argues that the variation in mtDNA within species or genera is way too small to represent the long time spans suggested by evolution. He doesn’t include selection in his analyses, though he could easily convince himself that this is a very real phenomenon. As in the example in humans above, if the ratio of synonymous vs. non-synonymous substitutions is compared between individuals with high vs. low homology, the difference leaves a strong signal of selection in mtDNA.
Jeanson uses mtDNA to track the speciation within a number of families and comes up with linear models of speciation in all the cases he evaluates. In his calculation of number of generations since the last common ancestor, Jeanson concludes that the time suggested by evolution is way too long to be accounted for by the differences within the species or genus in question. I have already pointed out that selection should be included in such calculations, but there is another objection that invalidates at least the non-human examples (fruit fly, water flea, and nematode), that is extinction. Jeanson makes no attempt to include extinction in his evaluation, though he appreciates that extinction is a very real phenomenon. This neglect leaves his analysis rather useless.
The first appearance in the fossil record of a genus or species need not be the last common ancestor of all modern members of that genus or species. That could appear much later if most other lines have become extinct.
Conclusion: Jeanson fails to account for the timing of speciation. If selection and extinction is taken in to account, the theory of evolution has no problem fitting the observations with the suggested time spans.
All in all, Jeanson fails on all four topics to show that creation can account for the observations, while the theory of evolution cannot.
Additional Points
Darwin didn’t know genetics
Jeanson postulates that because Darwin didn’t know genetics, there is good reason to question his theory. Let’s see if that is even relevant!
Species are defined by their traits! Traits are defined by genetics! Darwin didn’t know any genetics!
How then could Darwin write a book about the origin of species?
This seems like a legitimate question. But, consider the following premises of evolution:
If there are limited resources; If traits are inherited; If there is variation in inherited traits; If some source of new hereditary variation exists; If part of the inherited variation influence survival and, more importantly, reproduction. Then we can conclude the following:
Some hereditary traits, those that support reproduction more than the alternative traits, will spread in a population. Others will disappear. Hence, the combination of hereditary traits in the population will change over generations. This is evolution in its most basic form: Descent with modification.
Darwin’s bold idea was not only to suggest this process, but also to suggest that this process could go on forever, and that there is no limit to variation.
Evolution will take place whenever the premises are met, regardless of how heredity actually works. Knowledge of the details of heredity thus isn’t necessary to suggest evolution!
However, that doesn’t mean that knowledge of genetics is irrelevant. The details of genetics has to be such that variation actually is endless! It has to be such that brand new traits actually can occur!
When investigated, the genetic variation within and among species has to reflect the expectations that can be made from descent with modification. Otherwise, evolution would have been disproven.
That’s why genetics is such a relevant subject, when it comes to evolution.
Nevertheless, Jeanson’s assertion that Darwin’s theory was unfounded in his own times, fails.
Strange quotes
Here are a few strange quotes from the book:
Chapter 3:
“At the molecular level, major changes to the standard developmental pathway for vertebrae would be required to produce the giraffe’s signature structure [the long neck].”
Hardly. The neck of the giraffe is much longer than that of the okapi. They are both from the family Giraffidae, and therefore the same biblical ‘kind’ (Lightner 2012). Apparently, a factor 2 change in neck length is no problem within the biblical timescale of 4400 years.
Chapter 3:
“Could jellyfish become jaguars?”
Why such a silly example. No biologist has ever suggested this. Evolution can only result in transformations that can be linked by multiple small differences. The theory of evolution does not state that all organisms can evolve into any other.
Chapter 10:
“How can miracles have a place in science? If you’re an evolutionist, I would respond by pointing to Michael Behe’s published work.”
Michael Behe’s central ideas of ‘Intelligent design’ and ‘Irreducible complexity’ have never been accepted by ‘secular’ scientists. His books has been heavily criticized in ‘secular’ scientific journals. Referring to Behe as an example of a scientist who refers to miracles, only shows that Jeanson apparently is not aware of this fact.
Afterword:
“If atheistic evolution is true . . . How can you trust them [your eyes] to convey accurate information?”
If evolution is true all details in our anatomy are the result of mutation/selection through eons of time. Numerous small beneficial variations in our forefathers have been selected and added to beneficial variations in their ancestors. To what benefit would a variation in an eye be, if it didn’t help survival and, ultimately, reproduction? None! How can a variation in the eye help survival? Only by giving a slightly more accurate picture of the world. Therefore, according to evolution, we can trust our senses because they are the result of natural selection. That they are not 100% accurate is understandable. Random variation cannot be expected to result in perfection.
Jeanson is taking selection out of evolution, and then saying: Look, evolution doesn’t work.
I don’t know who first came up with this nonsense. I first came upon it when I heard Jason Lisle talk about his so-called ‘Ultimate Proof of Creation.’ It’s like taking God out Christianity and saying: Look, it doesn’t work.
A Short Concluding Remark
Would you trust an Atheist to teach your children about Christianity? If not—don’t trust a creationist to tell them about evolution!
References
Frello, S. 2017. ‘Reply to “Response to ‘On the Creationist View on mtDNA’.”’ Answers Research Journal 10: 237.
Jeanson, N. T. 2017a. Replacing Darwin: The New Origin of Species. Green Forest, Arkansas: Master Books.
Jeanson, N. T. 2017b. ‘Response to “Reply to ‘Response to ‘On the Creationist View on mtDNA’.”’ Answers Research Journal 10: 239–240.
Johnson, W. E., E. Eizirik, J. Pecon-Slattery, W. J. Murphy, A. Antunes, E. Teeling, and S. J. O’Brien. 2006. “The Late Miocene Radiation of Modern Felidae: A Genetic Assessment.” Science 311 (5757): 73–77.
Lightner, J. 2012. “Mammalian Ark Kinds.” Answers Research Journal 5: 151–204.
Prado-Martinez, J. et al. 2013. “Great Ape Genetic Diversity and Population History.” Nature 499 (7459): 471–475.