
Darwin at the Drugstore?
Testing the Biological Fitness of Antibiotic Resistant Bacteria
Keywords: Darwin, biology, antibiotic resistance, bacteria, evolution, macroevolution, microbiology, famine conditions, pathogen, growth rates
Introduction
The issue of antibiotic resistance in bacteria is growing. For example, methicillin-resistant Staphylococcus aureus (MRSA) is a leading human pathogen. New strains of MRSA are resistant to methicillin and numerous other antibiotics, placing the patient in significant danger. Indeed, the Centers for Disease Control revealed in 2007 that the number of deaths due to MRSA exceeded that of AIDS in the United States. It is no wonder that antibiotic resistance is one of the most significant and important health care issues. The ability of bacterial pathogens to evade traditional antibiotics is faster than the drug makers’ ability to make new types of clinical drugs (Gillen, 2007).
Evolutionary biologists have seized upon this rapid phenotypic change as alleged powerful evidence for Darwinian evolution. Thus, antibiotic resistance is an important two-fold issue involving empirical medical science and the volatile origins issue. It is frequently cited as a fact of evolution. Most recently, Carl Zimmer, in his new book, Microcosm: E. coli and the New Science of Life, uses drug resistance in Escherichia coli to bolster his arguments for macroevolution (or molecules-to-man evolution). One of his chapters, “Darwin at the Drugstore,” states that scientists are investigating drug resistance in E. coli to demonstrate their ability to undergo “rapid bursts of evolution,” (p. 97). Many biologists argue a case for Darwinian medicine—using evolutionary explanations to describe the wide variation in antibiotic resistance and adaption to new drugs.
Antibiotic resistance is certainly an example of change, but it is hardly a fact of macroevolution.
Antibiotic resistance is certainly an example of change, but it is hardly a fact of macroevolution (bacteria remain bacteria). Creation microbiologist, Dr. Kevin Anderson, states that such variation in bacteria is beneficial for their survival outcome in a clinical environment, but not a beneficial mutation. Anderson (2005) goes on to demonstrate how some “fitness” cost is often associated with mutations, although reversion mutations may eventually recover most, if not all, of this cost for some bacteria. A biological cost does occur in the loss of pre-existing cellular systems or functions. Such loss of cellular activity cannot legitimately be offered as a genetic means of demonstrating macroevolution.
Creation biologists have long been interested in discussing antibiotic resistance (Penrose, 1998) but have frequently cited other scientists’ data to make generalizations about drug-resistant bacteria being less fit overall than wild-type bacteria. Although this principle is theoretically simple to state on paper, experiments demonstrating fitness costs have been quite tricky in practice. In some cases, creation biologists have interpreted the data well; in other cases they have made wide, sweeping generalizations without data to back up their assertions.
Purpose of Study
In an on-going study at Liberty University, antibiotic resistance in the common red bacteria, Serratia marcescens, is being investigated to evaluate biological cost to bacteria due to resistance. The major objective of these experiments was to test the hypothesis that ampicillin-resistant bacteria are less “fit” than their wild-type counterparts. The investigation focused on the most common clinical, ampicillin-resistant S. marcescens strains (WFR) and compared with a wild-type strain. We evaluated the influence of ampicillin resistance on the bacteria’s overall fitness in a non-clinical environment.
Serratia marcescens is a Gram-negative, rod-shaped, facultative anaerobe (Figure 1), is ubiquitous in soil and water, and therefore is a common contaminant of food. It is easily grown in the lab and may serve as an ideal model for competition studies because of the natural color variation of S. marcescens (Gillen, 2008). In the wild, S. marcescens is noted for the production of a bright red pigment called prodigiosin (Williams, 1973). In contrast, 95% of clinical, resistant strains of Serratia are white or have little pigment (Von Graevenitz and Rubin, 1980).
S. marcescens is also a common cause of nosocomial (hospital) infections of the digestive and urinary tracts (Houdt et. al, 2007). In fact, 92% of all S. marcescens pathogens isolated from clinical settings have acquired ampicillin resistance (Graevenitz, 1980). The WF strain of Serratia marcescens was first isolated at the Texas Medical Center and has continued in the laboratory of Robert Williams for decades. This strain was chosen because it has characteristics like the vast majority of clinical strains (113/114 strains examined by Ding and Williams, 1983).
Figure 1. Phenotypic variation in Serratia marcescens can vary among red, pink, white, and other colors.
In our study, we compared the biological fitness of Serratia marcescens strains that are red, ampicillin-sensitive (AMPS) versus white, ampicillin resistant (AMPR). We wanted to ascertain whether antibiotic resistant mutants are less fit than wild-type. Fitness was defined by growth rate and colony “robustness” in minimal media.
Minimal Media Simulates Famine Conditions
Initially, it was difficult to demonstrate differences between wild-type and clinical strains in a rich media (Nutrient or Typticase-soy agar). There were no differences in growth rate or colony size. However, after switching to minimal media and observing hourly, the differences were readily observed. In order to confirm and extend the differences in growth rates between the sensitive BS303S strain (isolated from pond water) and the resistant WFR strain, a fitness/competition assay was performed. This assay sought to simulate famine conditions in the natural environment by utilizing minimal media and to evaluate the wild-type against ampicillin resistant, clinical strains exhibiting loss of prodigiosin production. Once subjected to conditions that were “harsh,” differences were seen in their performance (growth rate and robustness of colonies).
The Results Show a Difference
One analogy to explain the need to use minimal media in competition studies is the way in which weeds, not lush grass, grow during times of heat and drought. Like most people, we like a green lawn filled with robust, cultivated grass. During the spring when temperatures are moderate and plenty of rain comes, green grass grows well, along with a few weeds. However, during the summer, the maintenance of a good lawn is difficult: after a month of “drought,” clover and dandelions take over. The cultivated, green grass corresponds to antibiotic-resistant bacteria that have grown in the laboratory or (hospital-like environments), and the weeds represent the wild-type bacteria. In the spring, cultivated grasses, clover, and dandelions all grow nicely together producing a good lawn. When nutrients and proper temperature are maintained, both wild-type and resistant bacteria grow nicely in the Petri dish and in the body. But as conditions turn harsh, only the more fit bacteria (wild-type) grow well. Therefore, in order to simulate competition in the wild, bacteria must be grown on minimal media. Minimal media mimics better what bacteria experience in a natural environment over a period of time. This is the place where fitness can be accurately assessed. Given a rich media, they grow about the same (data not shown).
Sensitive and resistant S. marcescens show differences in growth rate. The greatest apparent differences observed were at 4 and 6 hours (Figure 2). When comparisons were made between wild-type and mutant strains at other times, differences in growth rate were not measurably different. The experiments suggest ampicillin resistance has a metabolic cost to the S. marcescens strains. At least in minimal media, this claim is true.
The largest differences in growth rates among Serratia strains (BS303S and WFR) were observed between 4 and 6 hours (Figure 2). BS303S after four hours had 51% and after six hours had 35% more growth than WFR. As incubation time increased, WFR caught up to BS303S, in terms of cell numbers. At the end of 10 hours post inoculation, there was only a 4% difference between BS303S and WFR. The wild-type BS303S also had larger colonies (data not shown). Thus, slower growth rates observed in the ampicillin-resistant, clinical strains compared to the ampicillin-sensitive, wild-type strains, demonstrate that due to this mutation, the bacteria experienced a decrease in net cellular function.
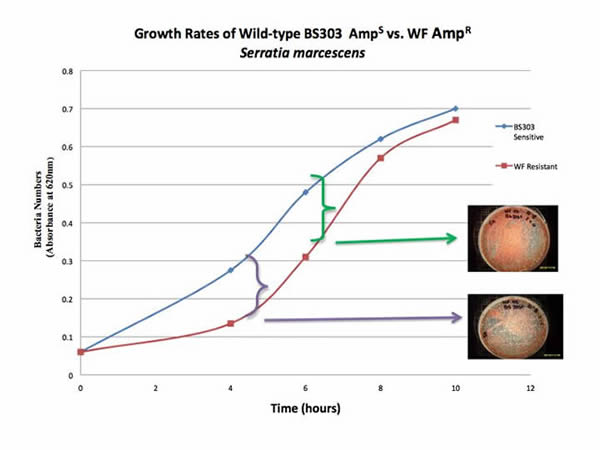
Figure 2. Comparisons of WFR (white, ampicillin-resistant) vs. BS 303 (red, wild-type). Note differences at 4 and 6 hours.
These results demonstrate that wild-type BS303S has a significant fitness advantage over the mutant strains due to its growth rate and colony size. Therefore, it can be argued that ampicillin resistance mutations reduce the growth rate and therefore the general biological fitness of S. marcescens. This study concurs with Anderson (2005) that while mutations providing antibiotic resistance may be beneficial in certain, specific environments, they often come at the expense of pre-existing function, and thus do not provide a mechanism for macroevolution.
It should be noted that we have found similar results with another Serratia marcescens clinical strain (933) and have observed it in E. coli. These findings will be reported in future papers. Our experience with competition studies reveals that demonstrating fitness costs in the laboratory is tricky and creation biologists should be careful about making dogmatic statements like, “Wild-type bacteria always outcompete antibiotic resistant mutants in nature.” Some bacteria that have an antibiotic resistance gene integrated into their chromosome can make compensatory mutations, over multiple, successive generations. These resistant bacteria compete favorably with wild-type bacteria in nature given certain environmental conditions (Criswell, 2004; 2007, personal communication).
Conclusions
Darwinian evolutionists attempt to offer explanations on antibiotic resistance and prescriptions for future drug development. If they simply suggest an awareness of on-going changes in pathogenic bacteria, we would concur. Bacteria do acquire resistance quickly, and many older drugs no longer work in hospitals and clinics. Creation microbiologists are interested in finding new drugs that will work and seek to heal the sick (Luke 10:9).
If evolutionists mean that one type of bacteria will “evolve” into another type, we would disagree based upon the evidence. A conclusion of these experiments is that acquiring antibiotic resistance through mutation or horizontal transfer is “costly.” There is a tradeoff: for survival in a clinical environment (i.e., antibiotics prevalent), there is a metabolic cost in terms of slower growth. This is called antagonistic pleiotropy. Antagonistic pleiotropy refers to the genetic expression of multiple competing effects, some beneficial, but others harmful to the organism. It often involves a case where reproduction and viability counter each other in biological fitness (Hedrick, 1999). The gene provides a benefit in one circumstance, but has cost in another.
The results of various experiments do show that bacteria can change quickly. Although the acquisition of antibiotic resistance does not demonstrate Darwinian evolution, it does demonstrate that bacteria were endowed by their Creator to change and adapt very quickly in an almost constantly changing environment. Bacteria have tremendous variability, yet there are limits to such change: the continuity, stability, and reliability of such bacteria are well known.
Bibliography
Kevin Anderson, “Is Bacterial Resistance to Antibiotics an Appropriate Example of Evolutionary Change?” Creation Research Society Quarterly 41 no. 4 (2005): 318.
D. Criswell, “The ‘Evolution’ of Antibiotic Resistance,” Acts & Facts 378 (2004), pp. 1–4.
Ming-Jer Ding and R.P. William, “Biosynthesis of Prodigiosin by White Strains of Serratia marcescens Isolated from Patients,” Journal of Clinical Microbiology (March 1983), pp. 476–480.
A.L. Gillen, The Genesis of Germs: Disease and the Coming Plagues in a Fallen World (Green Forest, Arkansas: Master Books, 2007).
A.L. Gillen, “Using Serratia marcescens as a Model to Elucidate Aspects of Germ Genesis in Bacteria,” in “Proceedings of the Microbe Forum, June 2007,” Creation Research Society Quarterly 45 no. 1 (2008): 240.
P.W. Hedrick, “Antagonistic Pleiotropy, and Genetic Polymorphism: A Perspective,” Heredity 82 (1999): 126–133.
Rob Van Houdt, Michael Givskov, and Chris Michiels, “Quorum Sensing in Serratia,” FEMS Microbiology 31 (2007): 407–424.
E. Penrose, “Bacterial Resistance to Antibiotics—a Case of Un-Natural Selection,” Creation Research Society Quarterly 35 (1998): 76–83.
A. Von Graevenitz and S.J. Rubin (eds.), The Genus Serratia (Boca Raton, FL: CRC Press, 1980).
R.P. Williams, “Biosynthesis of Prodigiosin, a Secondary Metabolite of Serratia marcescens,” Appl. Microbiol. 25 (1973): 396–402.
C. Zimmer, E. coli and the New Science of Life (New York: Pantheon Books, 2008).

Answers in Genesis is an apologetics ministry, dedicated to helping Christians defend their faith and proclaim the good news of Jesus Christ.
- Customer Service 800.778.3390
- Available Monday–Friday | 9 AM–5 PM ET
- © 2026 Answers in Genesis

